当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single Molecular Chelation Dynamics Reveals That DNA Has a Stronger Affinity toward Lead(II) than Cadmium(II)
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2022-02-23 , DOI: 10.1021/acs.jpcb.1c10487 Shu-Qian Hu 1 , Shi-Yong Ran 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2022-02-23 , DOI: 10.1021/acs.jpcb.1c10487 Shu-Qian Hu 1 , Shi-Yong Ran 1
Affiliation
|
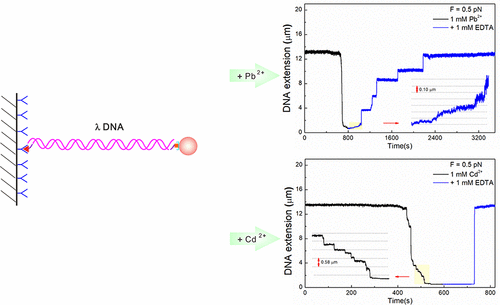
Lead ions can bind to DNA via nonelectrostatic interactions and hence alter its structure, which may be related to their adverse effects. The dynamics of Pb2+–DNA interaction has not been well understood. In this study, we report the monomolecular dynamics of the Pb2+–DNA interaction using a magnetic tweezers (MT) setup. We found that lead cations could induce DNA compaction at ionic strengths above 1 μM, which was also confirmed by morphology characterization. The chelation behavior of the Pb2+–DNA and the Cd2+–DNA complex solutions after adding EDTA were compared. The results showed that EDTA chelated with the bound metal ions on DNA and consequently led to restoring the DNA to its original length but with different restoration speeds for the two solutions. The fast binding dynamics and the slower chelation dynamics of the Pb2+ scenario compared to that of Cd2+ suggested that Pb2+ was more capable to induce DNA conformational change and that the Pb2+–DNA complex was more stable than the Cd2+–DNA complex. The stronger affinities for DNA bases and the inner binding of lead cations were two possible causes of the dynamics differences. Three agents, including EDTA, sodium gluconate, and SDBS, were used to remove the bound lead ions on DNA. It was shown that EDTA was the most efficient, and sodium gluconate could not fully restore DNA from its compact state. We concluded that both EDTA and SDBS were good candidates to restore the Pb2+-bound DNA to its original state.
中文翻译:

单分子螯合动力学揭示 DNA 对铅 (II) 的亲和力比对镉 (II) 的亲和力更强
铅离子可以通过非静电相互作用与 DNA 结合,从而改变其结构,这可能与其不利影响有关。Pb 2+ -DNA 相互作用的动力学尚未得到很好的理解。在这项研究中,我们使用磁性镊子 (MT) 装置报告了 Pb 2+ -DNA 相互作用的单分子动力学。我们发现铅阳离子可以在离子强度高于 1 μM 时诱导 DNA 压实,这也通过形态表征得到证实。Pb 2+ -DNA与Cd 2+的螯合行为– 比较添加 EDTA 后的 DNA 复合物溶液。结果表明,EDTA 与 DNA 上的结合金属离子螯合,从而导致 DNA 恢复到其原始长度,但两种溶液的恢复速度不同。与 Cd 2+相比, Pb 2+方案的快速结合动力学和较慢的螯合动力学表明 Pb 2+更能诱导 DNA 构象变化,并且 Pb 2+ -DNA复合物比 Cd 更稳定2+——DNA复合物。DNA碱基的更强亲和力和铅阳离子的内部结合是动力学差异的两个可能原因。三种试剂,包括 EDTA、葡萄糖酸钠和 SDBS,用于去除 DNA 上的结合铅离子。结果表明,EDTA 是最有效的,葡萄糖酸钠不能完全恢复 DNA 的紧致状态。我们得出结论,EDTA 和 SDBS 都是将 Pb 2+结合的DNA 恢复到其原始状态的良好候选者。
更新日期:2022-02-23
中文翻译:

单分子螯合动力学揭示 DNA 对铅 (II) 的亲和力比对镉 (II) 的亲和力更强
铅离子可以通过非静电相互作用与 DNA 结合,从而改变其结构,这可能与其不利影响有关。Pb 2+ -DNA 相互作用的动力学尚未得到很好的理解。在这项研究中,我们使用磁性镊子 (MT) 装置报告了 Pb 2+ -DNA 相互作用的单分子动力学。我们发现铅阳离子可以在离子强度高于 1 μM 时诱导 DNA 压实,这也通过形态表征得到证实。Pb 2+ -DNA与Cd 2+的螯合行为– 比较添加 EDTA 后的 DNA 复合物溶液。结果表明,EDTA 与 DNA 上的结合金属离子螯合,从而导致 DNA 恢复到其原始长度,但两种溶液的恢复速度不同。与 Cd 2+相比, Pb 2+方案的快速结合动力学和较慢的螯合动力学表明 Pb 2+更能诱导 DNA 构象变化,并且 Pb 2+ -DNA复合物比 Cd 更稳定2+——DNA复合物。DNA碱基的更强亲和力和铅阳离子的内部结合是动力学差异的两个可能原因。三种试剂,包括 EDTA、葡萄糖酸钠和 SDBS,用于去除 DNA 上的结合铅离子。结果表明,EDTA 是最有效的,葡萄糖酸钠不能完全恢复 DNA 的紧致状态。我们得出结论,EDTA 和 SDBS 都是将 Pb 2+结合的DNA 恢复到其原始状态的良好候选者。

































 京公网安备 11010802027423号
京公网安备 11010802027423号