当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-02-10 , DOI: 10.1021/jacs.1c08402 Andreas Luttens 1 , Hjalmar Gullberg 2 , Eldar Abdurakhmanov 3 , Duy Duc Vo 1 , Dario Akaberi 4 , Vladimir O Talibov 5 , Natalia Nekhotiaeva 2 , Laura Vangeel 6, 7 , Steven De Jonghe 6, 7 , Dirk Jochmans 6, 7 , Janina Krambrich 4 , Ali Tas 8 , Bo Lundgren 2 , Ylva Gravenfors 9 , Alexander J Craig 10 , Yoseph Atilaw 11 , Anja Sandström 10 , Lindon W K Moodie 10, 12 , Åke Lundkvist 4 , Martijn J van Hemert 8 , Johan Neyts 6, 7 , Johan Lennerstrand 13 , Jan Kihlberg 11 , Kristian Sandberg 10, 14, 15 , U Helena Danielson 3 , Jens Carlsson 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-02-10 , DOI: 10.1021/jacs.1c08402 Andreas Luttens 1 , Hjalmar Gullberg 2 , Eldar Abdurakhmanov 3 , Duy Duc Vo 1 , Dario Akaberi 4 , Vladimir O Talibov 5 , Natalia Nekhotiaeva 2 , Laura Vangeel 6, 7 , Steven De Jonghe 6, 7 , Dirk Jochmans 6, 7 , Janina Krambrich 4 , Ali Tas 8 , Bo Lundgren 2 , Ylva Gravenfors 9 , Alexander J Craig 10 , Yoseph Atilaw 11 , Anja Sandström 10 , Lindon W K Moodie 10, 12 , Åke Lundkvist 4 , Martijn J van Hemert 8 , Johan Neyts 6, 7 , Johan Lennerstrand 13 , Jan Kihlberg 11 , Kristian Sandberg 10, 14, 15 , U Helena Danielson 3 , Jens Carlsson 1
Affiliation
|
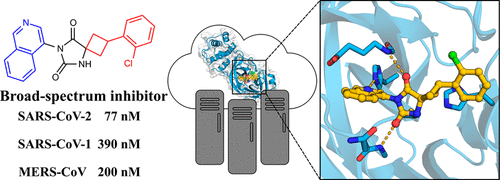
Drugs targeting SARS-CoV-2 could have saved millions of lives during the COVID-19 pandemic, and it is now crucial to develop inhibitors of coronavirus replication in preparation for future outbreaks. We explored two virtual screening strategies to find inhibitors of the SARS-CoV-2 main protease in ultralarge chemical libraries. First, structure-based docking was used to screen a diverse library of 235 million virtual compounds against the active site. One hundred top-ranked compounds were tested in binding and enzymatic assays. Second, a fragment discovered by crystallographic screening was optimized guided by docking of millions of elaborated molecules and experimental testing of 93 compounds. Three inhibitors were identified in the first library screen, and five of the selected fragment elaborations showed inhibitory effects. Crystal structures of target–inhibitor complexes confirmed docking predictions and guided hit-to-lead optimization, resulting in a noncovalent main protease inhibitor with nanomolar affinity, a promising in vitro pharmacokinetic profile, and broad-spectrum antiviral effect in infected cells.
中文翻译:

超大型虚拟筛选可识别具有广谱抗冠状病毒活性的 SARS-CoV-2 主要蛋白酶抑制剂
在 COVID-19 大流行期间,针对 SARS-CoV-2 的药物可能挽救了数百万人的生命,现在开发冠状病毒复制抑制剂以应对未来的爆发至关重要。我们探索了两种虚拟筛选策略,以在超大型化学文库中寻找 SARS-CoV-2 主要蛋白酶的抑制剂。首先,基于结构的对接被用于筛选针对活性位点的 2.35 亿个虚拟化合物的多样化库。一百个排名靠前的化合物在结合和酶分析中进行了测试。其次,通过对数百万个精细分子的对接和93种化合物的实验测试,对晶体筛选发现的片段进行了优化。在第一个文库筛选中确定了三种抑制剂,其中五个选定的片段精化显示出抑制作用。
更新日期:2022-02-10
中文翻译:

超大型虚拟筛选可识别具有广谱抗冠状病毒活性的 SARS-CoV-2 主要蛋白酶抑制剂
在 COVID-19 大流行期间,针对 SARS-CoV-2 的药物可能挽救了数百万人的生命,现在开发冠状病毒复制抑制剂以应对未来的爆发至关重要。我们探索了两种虚拟筛选策略,以在超大型化学文库中寻找 SARS-CoV-2 主要蛋白酶的抑制剂。首先,基于结构的对接被用于筛选针对活性位点的 2.35 亿个虚拟化合物的多样化库。一百个排名靠前的化合物在结合和酶分析中进行了测试。其次,通过对数百万个精细分子的对接和93种化合物的实验测试,对晶体筛选发现的片段进行了优化。在第一个文库筛选中确定了三种抑制剂,其中五个选定的片段精化显示出抑制作用。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号