当前位置:
X-MOL 学术
›
Chemosphere
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploration of binding mechanism of triclosan towards cancer markers using molecular docking and molecular dynamics
Chemosphere ( IF 8.1 ) Pub Date : 2022-01-06 , DOI: 10.1016/j.chemosphere.2022.133550 Prashant Bhardwaj 1 , G P Biswas 2 , Nibedita Mahata 3 , Susanta Ghanta 4 , Biswanath Bhunia 5
Chemosphere ( IF 8.1 ) Pub Date : 2022-01-06 , DOI: 10.1016/j.chemosphere.2022.133550 Prashant Bhardwaj 1 , G P Biswas 2 , Nibedita Mahata 3 , Susanta Ghanta 4 , Biswanath Bhunia 5
Affiliation
|
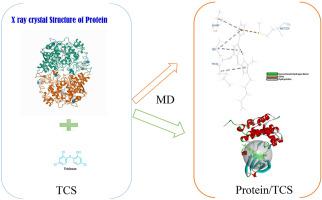
The molecule 5-chloro-2-(2,4-dichlorophenoxy) phenol is well-known as Triclosan (TCS), which is also a potential endocrine disrupting synthetic chemical. TCS exposure has been connected to the control of the human enoyl-acyl carrier protein-reductase (hER), which has been linked to a range of life threatening diseases. However, other than hER, the new protein targets for TCS that are responsible for a variety of cancers are yet unclear. The goal of this work is to investigate into the protein binding patterns of TCS and proteins from various cancer signaling pathways. Discovery Studio 4.1 was used to perform molecular docking and molecular dynamics (MD) on the protein-triclosan complex. The proteins were first screened using CHARMM-based docking with a CDOCKER energy greater than −21.40 kcal/mol. The CDOCKER energies of Fas-associated death domain (FADD), Receptor-interacting protein 1 (RIP1), F-κB-inducing kinase (NIK), c-Jun N -terminal kinase (JNK), Apoptosis signal-regulating kinase 1 (ASK1), B-cell lymphoma 2 (Bcl-2), Apoptosis-inducing factor (AIF), α-tubulin, and Actin were −20.68 kcal/mol, −26.88 kcal/mol, −23.43 kcal/mol, −22.21 kcal/mol, −20.40 kcal/mol, −21.10 kcal/mol, −20.98 kcal/mol, −24.67 kcal/mol, and −23.09 kcal/mol respectively. MD was performed on the screened proteins by standard dynamics cascade tool using CHARMM Force field. The MD results were accessed using the energy–time graph, root-mean-square deviation (RMSD), and root mean square fluctuations (RMSF). The 100 conformers of α-tubulin, NIK, FADD, and RIP1 were found to have a trend of increasing RMSD, whereas Bcl-2, ASK1, AIF, Actin, and JNK proteins had lower RMSD values. In compared to FADD, AIF, and JNK, the RMSF variations of the Bcl-2, ASK1, α-tubulin, Actin, NIK, and RIP1 residues were shown to be high. Similar patterns were seen in the energy variations, which range from 1000 kcal/mol to 2000 kcal/mol. RIP1 and Bcl-2 showed more variation in the sidechain RMSF in comparison to FADD, ASK1, AIF, Actin, α-tubulin, NIK and JNK. Thus, it can be postulated that AIF and JNK proteins of apoptosis signaling pathway are pivotal in the TCS mediated reactions.
中文翻译:

利用分子对接和分子动力学探索三氯生对癌症标志物的结合机制
分子 5-氯-2-(2,4-二氯苯氧基)苯酚是众所周知的三氯生 (TCS),它也是一种潜在的内分泌干扰合成化学物质。TCS 暴露与人烯酰基载体蛋白还原酶 (hER) 的控制有关,而 hER与一系列危及生命的疾病有关。然而,除了 hER 之外,导致多种癌症的 TCS 的新蛋白靶点尚不清楚。这项工作的目标是研究 TCS 的蛋白质结合模式和来自各种癌症信号通路的蛋白质。Discovery Studio 4.1 用于对蛋白质-三氯生复合物进行分子对接和分子动力学 (MD)。首先使用基于 CHARMM 的对接筛选蛋白质,CDOCKER 能量大于 -21.40 kcal/mol。Fas 相关死亡结构域 (FADD)、受体相互作用蛋白 1 (RIP1)、F-κB 诱导激酶 (NIK)、c-Jun N 末端激酶 (JNK)、细胞凋亡信号调节激酶 1 (ASK1)、B 细胞淋巴瘤 2 (Bcl-2)、凋亡诱导因子 (AIF)、α-微管蛋白和肌动蛋白的 CDOCKER 能量分别为 -20.68 kcal/mol、-26.88 kcal/mol、-23.43 kcal/mol、-22.21 kcal/mol、-20.40 kcal/mol、-21.10 kcal/mol、-20.98 kcal/mol、 分别为 -24.67 kcal/mol 和 -23.09 kcal/mol。使用 CHARMM 力场通过标准动力学级联工具对筛选的蛋白质进行 MD。使用能量-时间图、均方根偏差 (RMSD) 和均方根波动 (RMSF) 获取 MD 结果。发现 α-tubulin 、 NIK 、 FADD 和 RIP1 的 100 个构象子具有增加 RMSD 的趋势,而 Bcl-2 、 ASK1 、 AIF 、 Actin 和 JNK 蛋白的 RMSD 值较低。 与 FADD 、 AIF 和 JNK 相比,Bcl-2 、 ASK1 、 α-微管蛋白、肌动蛋白、 NIK 和 RIP1 残基的 RMSF 变异显示很高。在能量变化中也观察到类似的模式,范围从 1000 kcal/mol 到 2000 kcal/mol。与 FADD、ASK1、AIF、Actin、α-tubulin、NIK 和 JNK 相比,RIP1 和 Bcl-2 在侧链 RMSF 中显示出更多变化。因此,可以假设细胞凋亡信号通路的 AIF 和 JNK 蛋白在 TCS 介导的反应中起关键作用。
更新日期:2022-01-06
中文翻译:

利用分子对接和分子动力学探索三氯生对癌症标志物的结合机制
分子 5-氯-2-(2,4-二氯苯氧基)苯酚是众所周知的三氯生 (TCS),它也是一种潜在的内分泌干扰合成化学物质。TCS 暴露与人烯酰基载体蛋白还原酶 (hER) 的控制有关,而 hER与一系列危及生命的疾病有关。然而,除了 hER 之外,导致多种癌症的 TCS 的新蛋白靶点尚不清楚。这项工作的目标是研究 TCS 的蛋白质结合模式和来自各种癌症信号通路的蛋白质。Discovery Studio 4.1 用于对蛋白质-三氯生复合物进行分子对接和分子动力学 (MD)。首先使用基于 CHARMM 的对接筛选蛋白质,CDOCKER 能量大于 -21.40 kcal/mol。Fas 相关死亡结构域 (FADD)、受体相互作用蛋白 1 (RIP1)、F-κB 诱导激酶 (NIK)、c-Jun N 末端激酶 (JNK)、细胞凋亡信号调节激酶 1 (ASK1)、B 细胞淋巴瘤 2 (Bcl-2)、凋亡诱导因子 (AIF)、α-微管蛋白和肌动蛋白的 CDOCKER 能量分别为 -20.68 kcal/mol、-26.88 kcal/mol、-23.43 kcal/mol、-22.21 kcal/mol、-20.40 kcal/mol、-21.10 kcal/mol、-20.98 kcal/mol、 分别为 -24.67 kcal/mol 和 -23.09 kcal/mol。使用 CHARMM 力场通过标准动力学级联工具对筛选的蛋白质进行 MD。使用能量-时间图、均方根偏差 (RMSD) 和均方根波动 (RMSF) 获取 MD 结果。发现 α-tubulin 、 NIK 、 FADD 和 RIP1 的 100 个构象子具有增加 RMSD 的趋势,而 Bcl-2 、 ASK1 、 AIF 、 Actin 和 JNK 蛋白的 RMSD 值较低。 与 FADD 、 AIF 和 JNK 相比,Bcl-2 、 ASK1 、 α-微管蛋白、肌动蛋白、 NIK 和 RIP1 残基的 RMSF 变异显示很高。在能量变化中也观察到类似的模式,范围从 1000 kcal/mol 到 2000 kcal/mol。与 FADD、ASK1、AIF、Actin、α-tubulin、NIK 和 JNK 相比,RIP1 和 Bcl-2 在侧链 RMSF 中显示出更多变化。因此,可以假设细胞凋亡信号通路的 AIF 和 JNK 蛋白在 TCS 介导的反应中起关键作用。































 京公网安备 11010802027423号
京公网安备 11010802027423号