Journal of Molecular Structure ( IF 4.0 ) Pub Date : 2021-12-24 , DOI: 10.1016/j.molstruc.2021.132258 Uddipan Bhattacharya 1 , Saroj Kumar Panda 1 , Parth Sarthi Sen Gupta 1 , Malay Kumar Rana 1
|
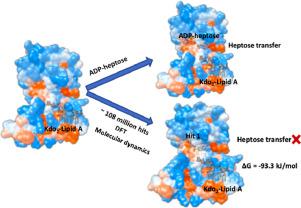
Heptosyltransferase I (HEP I) belongs to the broad family of glycosyltransferase-B (GT-B) enzymes, which transfers heptose sugar necessary for the lipopolysaccharide (LPS) synthesis. LPS is a significant component of the external cell membrane of Gram-negative bacteria, e.g., Escherichia coli. Interruption of heptose sugar addition during LPS biosynthesis greatly reduces bacterial infections and antibiotic resistance. Thus, inhibition of HEP I is indispensable to counteract this problem. Here, we have performed virtual screening employing structure-based pharmacophore, docking, pharmacokinetics, and electronic descriptors to identify four important hit molecules from a pool of ∼108 million bioactive molecules existing in ZINC, PubChem, and CHEMBL databases against HEP I. Pharmacokinetics and all electronic descriptors describing the bioavailability of the selected hits fall within the permissible limits. Further, molecular dynamics simulations of the shortlisted hits along with a reference molecule discern the conformational stability of the bound complexes, characterized by RMSD, RMSF, Rg, SASA, and PCA. The large negative MM-PBSA-based binding free energy validates the initial screening results and hence the inhibitory potential of the hits. The study suggests that the best three candidates, hit 1 (CHEMBL1438912), hit 3 (CHEMBL13895), and hit 4 (CHEMBL320880), can be very promising to retard the heptose sugar transfer and eradicate the antibiotic resistance of E. coli, providing molecular insights into the inhibition of HEP I. This may facilitate further in-vitro studies of the best hits and targeted antibacterial drug development.
中文翻译:

Heptosyltransferase I 的抑制剂防止庚糖转移对抗大肠杆菌的抗生素耐药性:通过 DFT 和分子动力学进行能量学和稳定性分析
庚糖基转移酶 I (HEP I) 属于糖基转移酶-B (GT-B) 酶的广泛家族,可转移脂多糖 (LPS) 合成所需的庚糖。LPS 是革兰氏阴性菌(如大肠杆菌)外细胞膜的重要成分. LPS 生物合成过程中庚糖添加的中断大大减少了细菌感染和抗生素耐药性。因此,HEP I 的抑制对于解决这个问题是必不可少的。在这里,我们使用基于结构的药效团、对接、药代动力学和电子描述符进行了虚拟筛选,以从 ZINC、PubChem 和 CHEMBL 数据库中存在的约 1.08 亿个生物活性分子中针对 HEP I 识别出四种重要的命中分子。所有描述所选命中生物利用度的电子描述符都在允许的范围内。此外,入围命中和参考分子的分子动力学模拟辨别结合复合物的构象稳定性,特征在于 RMSD、RMSF、R g、SASA 和 PCA。基于 MM-PBSA 的大负结合自由能验证了初始筛选结果,因此验证了命中的抑制潜力。该研究表明,命中 1 (CHEMBL1438912)、命中 3 (CHEMBL13895) 和命中 4 (CHEMBL320880) 的最佳三个候选药物可以非常有希望延缓庚糖转移并根除大肠杆菌的抗生素耐药性,提供分子深入了解 HEP I 的抑制作用。这可能有助于对最佳命中和靶向抗菌药物开发的进一步体外研究。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号