当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Coordinated Actions of Cas9 HNH and RuvC Nuclease Domains Are Regulated by the Bridge Helix and the Target DNA Sequence
Biochemistry ( IF 2.9 ) Pub Date : 2021-11-10 , DOI: 10.1021/acs.biochem.1c00354
Kesavan Babu 1 , Venkatesan Kathiresan 2 , Pratibha Kumari 3 , Sydney Newsom 1 , Hari Priya Parameshwaran 1 , Xiongping Chen 3 , Jin Liu 3 , Peter Z Qin 2 , Rakhi Rajan 1
Biochemistry ( IF 2.9 ) Pub Date : 2021-11-10 , DOI: 10.1021/acs.biochem.1c00354
Kesavan Babu 1 , Venkatesan Kathiresan 2 , Pratibha Kumari 3 , Sydney Newsom 1 , Hari Priya Parameshwaran 1 , Xiongping Chen 3 , Jin Liu 3 , Peter Z Qin 2 , Rakhi Rajan 1
Affiliation
|
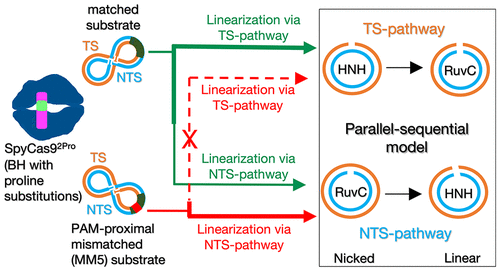
CRISPR-Cas systems are RNA-guided nucleases that provide adaptive immune protection in bacteria and archaea against intruding genomic materials. Cas9, a type-II CRISPR effector protein, is widely used for gene editing applications since a single guide RNA can direct Cas9 to cleave specific genomic targets. The conformational changes associated with RNA/DNA binding are being modulated to develop Cas9 variants with reduced off-target cleavage. Previously, we showed that proline substitutions in the arginine-rich bridge helix (BH) of Streptococcus pyogenes Cas9 (SpyCas9-L64P-K65P, SpyCas92Pro) improve target DNA cleavage selectivity. In this study, we establish that kinetic analysis of the cleavage of supercoiled plasmid substrates provides a facile means to analyze the use of two parallel routes for DNA linearization by SpyCas9: (i) nicking by HNH followed by RuvC cleavage (the TS (target strand) pathway) and (ii) nicking by RuvC followed by HNH cleavage (the NTS (nontarget strand) pathway). BH substitutions and DNA mismatches alter the individual rate constants, resulting in changes in the relative use of the two pathways and the production of nicked and linear species within a given pathway. The results reveal coordinated actions between HNH and RuvC to linearize DNA, which is modulated by the integrity of the BH and the position of the mismatch in the substrate, with each condition producing distinct conformational energy landscapes as observed by molecular dynamics simulations. Overall, our results indicate that BH interactions with RNA/DNA enable target DNA discrimination through the differential use of the parallel sequential pathways driven by HNH/RuvC coordination.
中文翻译:

Cas9 HNH 和 RuvC 核酸酶结构域的协调作用受桥螺旋和目标 DNA 序列的调节
CRISPR-Cas 系统是 RNA 引导的核酸酶,可为细菌和古细菌提供适应性免疫保护,防止基因组物质入侵。 Cas9 是一种 II 型 CRISPR 效应蛋白,广泛用于基因编辑应用,因为单个引导 RNA 可以指导 Cas9 切割特定的基因组靶标。与 RNA/DNA 结合相关的构象变化正在被调节,以开发具有减少脱靶切割的 Cas9 变体。之前,我们发现化脓性链球菌Cas9 (SpyCas9-L64P-K65P、SpyCas9 2Pro ) 富含精氨酸的桥螺旋 (BH) 中的脯氨酸取代可提高靶标 DNA 切割选择性。在本研究中,我们确定超螺旋质粒底物裂解的动力学分析提供了一种简便的方法来分析 SpyCas9 对 DNA 线性化的两条平行途径的使用:(i) HNH 切口,然后是 RuvC 裂解(TS(目标链) ) 途径) 和 (ii) RuvC 产生切口,然后进行 HNH 切割(NTS(非目标链)途径)。 BH 取代和 DNA 错配会改变各个速率常数,从而导致两条途径的相对使用以及给定途径内切口和线性物种的产生发生变化。结果揭示了 HNH 和 RuvC 之间协调作用以线性化 DNA,这是由 BH 的完整性和底物中错配的位置调节的,每个条件都会产生不同的构象能量景观,如分子动力学模拟所观察到的。总体而言,我们的结果表明,BH 与 RNA/DNA 的相互作用能够通过 HNH/RuvC 协调驱动的并行顺序途径的差异使用来区分目标 DNA。
更新日期:2021-12-14
中文翻译:

Cas9 HNH 和 RuvC 核酸酶结构域的协调作用受桥螺旋和目标 DNA 序列的调节
CRISPR-Cas 系统是 RNA 引导的核酸酶,可为细菌和古细菌提供适应性免疫保护,防止基因组物质入侵。 Cas9 是一种 II 型 CRISPR 效应蛋白,广泛用于基因编辑应用,因为单个引导 RNA 可以指导 Cas9 切割特定的基因组靶标。与 RNA/DNA 结合相关的构象变化正在被调节,以开发具有减少脱靶切割的 Cas9 变体。之前,我们发现化脓性链球菌Cas9 (SpyCas9-L64P-K65P、SpyCas9 2Pro ) 富含精氨酸的桥螺旋 (BH) 中的脯氨酸取代可提高靶标 DNA 切割选择性。在本研究中,我们确定超螺旋质粒底物裂解的动力学分析提供了一种简便的方法来分析 SpyCas9 对 DNA 线性化的两条平行途径的使用:(i) HNH 切口,然后是 RuvC 裂解(TS(目标链) ) 途径) 和 (ii) RuvC 产生切口,然后进行 HNH 切割(NTS(非目标链)途径)。 BH 取代和 DNA 错配会改变各个速率常数,从而导致两条途径的相对使用以及给定途径内切口和线性物种的产生发生变化。结果揭示了 HNH 和 RuvC 之间协调作用以线性化 DNA,这是由 BH 的完整性和底物中错配的位置调节的,每个条件都会产生不同的构象能量景观,如分子动力学模拟所观察到的。总体而言,我们的结果表明,BH 与 RNA/DNA 的相互作用能够通过 HNH/RuvC 协调驱动的并行顺序途径的差异使用来区分目标 DNA。































 京公网安备 11010802027423号
京公网安备 11010802027423号