Molecular Therapy ( IF 12.1 ) Pub Date : 2021-11-02 , DOI: 10.1016/j.ymthe.2021.10.026
Friederike Knipping 1 , Gregory A Newby 2 , Cindy R Eide 1 , Amber N McElroy 1 , Sarah C Nielsen 1 , Kyle Smith 1 , Yongxing Fang 3 , Tatjana I Cornu 4 , Caroline Costa 5 , Alejandra Gutierrez-Guerrero 5 , Samuel P Bingea 1 , Colby J Feser 1 , Benjamin Steinbeck 1 , Keli L Hippen 1 , Bruce R Blazar 1 , Anton McCaffrey 6 , Claudio Mussolino 4 , Els Verhoeyen 7 , Jakub Tolar 1 , David R Liu 2 , Mark J Osborn 1
|
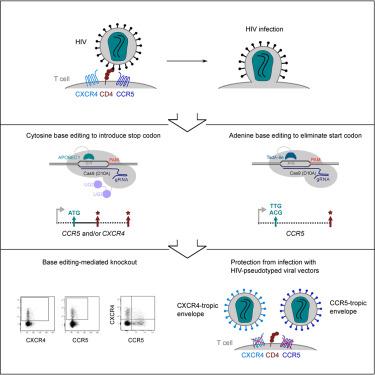
Disruption of CCR5 or CXCR4, the main human immunodeficiency virus type 1 (HIV-1) co-receptors, has been shown to protect primary human CD4+ T cells from HIV-1 infection. Base editing can install targeted point mutations in cellular genomes, and can thus efficiently inactivate genes by introducing stop codons or eliminating start codons without double-stranded DNA break formation. Here, we applied base editors for individual and simultaneous disruption of both co-receptors in primary human CD4+ T cells. Using cytosine base editors we observed premature stop codon introduction in up to 89% of sequenced CCR5 or CXCR4 alleles. Using adenine base editors we eliminated the start codon in CCR5 in up to 95% of primary human CD4+ T cell and up to 88% of CD34+ hematopoietic stem and progenitor cell target alleles. Genome-wide specificity analysis revealed low numbers of off-target mutations that were introduced by base editing, located predominantly in intergenic or intronic regions. We show that our editing strategies prevent transduction with CCR5-tropic and CXCR4-tropic viral vectors in up to 79% and 88% of human CD4+ T cells, respectively. The engineered T cells maintained functionality and overall our results demonstrate the effectiveness of base-editing strategies for efficient and specific ablation of HIV co-receptors in clinically relevant cell types.
中文翻译:

使用碱基编辑破坏原代人 T 细胞和造血干细胞和祖细胞中的 HIV-1 共受体 CCR5 和 CXCR4
破坏主要的人类免疫缺陷病毒 1 型 (HIV-1) 辅助受体 CCR5 或 CXCR4 已被证明可以保护原代人类 CD4 + T 细胞免受 HIV-1 感染。碱基编辑可以在细胞基因组中安装靶向点突变,因此可以通过引入终止密码子或消除起始密码子而不形成双链 DNA 断裂来有效地使基因失活。在这里,我们应用碱基编辑器单独和同时破坏原代人 CD4 + T 细胞中的两种共受体。使用胞嘧啶碱基编辑器,我们观察到在多达 89% 的已测序CCR5或CXCR4等位基因中过早引入终止密码子。使用腺嘌呤碱基编辑器,我们消除了CCR5中的起始密码子在高达 95% 的原代人 CD4 + T 细胞和高达 88% 的 CD34 +造血干细胞和祖细胞靶等位基因中。全基因组特异性分析显示,碱基编辑引入的脱靶突变数量很少,主要位于基因间或内含子区域。我们表明,我们的编辑策略分别阻止了高达 79% 和 88% 的人类 CD4 + T 细胞中的 CCR5 嗜性和 CXCR4 嗜性病毒载体转导。工程化的 T 细胞保持了功能,总体而言,我们的结果证明了碱基编辑策略在临床相关细胞类型中有效和特异性消融 HIV 共受体的有效性。

































 京公网安备 11010802027423号
京公网安备 11010802027423号