Molecular Phylogenetics and Evolution ( IF 3.6 ) Pub Date : 2021-08-05 , DOI: 10.1016/j.ympev.2021.107287 Luiz Henrique M Fonseca 1
|
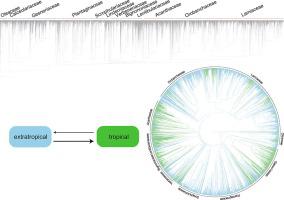
Lamiales is one of the most intractable orders of flowering plants, with several changes in family composition, and circumscription throughout history. The order is worldwide distributed, occurring in tropical forests and frozen habitats. In this study, a comprehensive phylogeny of Lamiales was reconstructed using DNA sequences. The tree was used to infer dispersal patterns, focusing on the tropics and extratropics. Molecular and species geographic data available from public repositories were combined to address both objectives. A total of 6,910 species, and 842 genera of Lamiales were sampled using the Python tool PyPHLAWD. The tree was inferred using RAxML, and recovered a monophyletic Lamiales. All 26 families were recovered as monophyletic with high support. The families Bignoniaceae, and Plantaginaceae are remarkable examples. The first emerged as monophyletic and included tribe Jacarandeae, while the later emerged as monophyletic in its sensu lato and included both the tribes Angelonieae, and Gratioleae. Distribution points for all species were retrieved from GBIF. After filtering, 1,136,425 records were retained. Species were coded as present in extratropical or tropical environments. The in and out of the tropics dispersal patterns were inferred using a maximum likelihood approach that identifies hidden rate changes. The model recovered higher rates of transition from extratropics to tropics, estimating two rates of state transitions. When ancestral states are considered, more discrete transitions from extratropics to tropics were observed. The extratropical state was also inferred for the crown node of Lamiales and old nested nodes, revealing a rare pattern of transitions to the tropics throughout the upper Cretaceous and Tertiary. A significant phylogenetic signal was recovered for the in and out of the tropics dispersal patterns, showing that state transitions are not frequent enough to erase the effect of tree structure on the data.
中文翻译:

结合分子和地理数据来推断 Lamiales 的系统发育及其在热带内外的扩散模式
Laamiales 是开花植物中最难处理的目之一,在家族组成和历史上都有一些变化。该目分布于世界各地,发生在热带森林和冰冻栖息地。在这项研究中,使用 DNA 序列重建了 Laamiales 的全面系统发育。这棵树被用来推断扩散模式,重点关注热带和外热带。将公共存储库中可用的分子和物种地理数据结合起来以实现这两个目标。使用 Python 工具 PyPHLAWD 对总共 6,910 个物种和 842 个属的 Lamiales 进行了采样。这棵树是使用 RAxML 推断的,并恢复了一个单系的 Lamiales。所有 26 个家庭都被恢复为具有高度支持的单系。Bignoniaceae 和 Plantaginaceae 是显着的例子。sensu lato包括 Angelonieae 和 Gratioleae 两个部落。从 GBIF 检索到所有物种的分布点。过滤后,保留了 1,136,425 条记录。物种被编码为存在于温带或热带环境中。使用识别隐藏速率变化的最大似然方法推断出热带扩散模式的进出。该模型恢复了从外热带到热带的更高转变率,估计了两种状态转变率。当考虑祖先状态时,观察到从外热带到热带的更离散的转变。温带状态也被推断为 Lamiales 的冠节点和旧的嵌套节点,揭示了在整个上白垩纪和第三纪向热带过渡的罕见模式。对于进出热带的扩散模式,恢复了重要的系统发育信号,































 京公网安备 11010802027423号
京公网安备 11010802027423号