Molecular Cell ( IF 14.5 ) Pub Date : 2021-06-30 , DOI: 10.1016/j.molcel.2021.06.008 Erica S M Vos 1 , Christian Valdes-Quezada 1 , Yike Huang 1 , Amin Allahyar 1 , Marjon J A M Verstegen 1 , Anna-Karina Felder 1 , Floor van der Vegt 1 , Esther C H Uijttewaal 1 , Peter H L Krijger 1 , Wouter de Laat 1
|
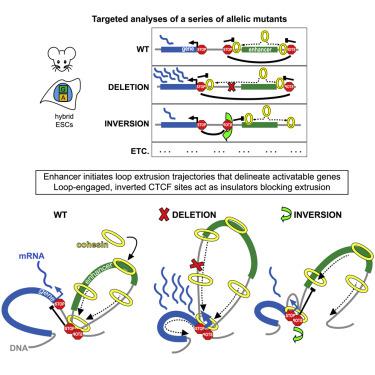
To understand how chromatin domains coordinate gene expression, we dissected select genetic elements organizing topology and transcription around the Prdm14 super enhancer in mouse embryonic stem cells. Taking advantage of allelic polymorphisms, we developed methods to sensitively analyze changes in chromatin topology, gene expression, and protein recruitment. We show that enhancer insulation does not rely strictly on loop formation between its flanking boundaries, that the enhancer activates the Slco5a1 gene beyond its prominent domain boundary, and that it recruits cohesin for loop extrusion. Upon boundary inversion, we find that oppositely oriented CTCF terminates extrusion trajectories but does not stall cohesin, while deleted or mutated CTCF sites allow cohesin to extend its trajectory. Enhancer-mediated gene activation occurs independent of paused loop extrusion near the gene promoter. We expand upon the loop extrusion model to propose that cohesin loading and extrusion trajectories originating at an enhancer contribute to gene activation.
中文翻译:

CTCF 边界和超级增强子之间的相互作用控制粘连蛋白挤出轨迹和基因表达
为了了解染色质结构域如何协调基因表达,我们剖析了小鼠胚胎干细胞中Prdm14超级增强子周围组织拓扑和转录的选定遗传元件。利用等位基因多态性,我们开发了灵敏分析染色质拓扑、基因表达和蛋白质招募变化的方法。我们表明,增强子绝缘并不严格依赖于其侧翼边界之间的环形成,增强子激活超出其突出结构域边界的Slco5a1基因,并且它招募粘连蛋白以进行环挤出。在边界反转后,我们发现相反方向的 CTCF 终止挤出轨迹,但不会阻止粘连蛋白,而删除或突变的 CTCF 位点允许粘连蛋白延长其轨迹。增强子介导的基因激活独立于基因启动子附近暂停的环挤出而发生。我们扩展了环挤出模型,提出源自增强子的粘连蛋白加载和挤出轨迹有助于基因激活。













































 京公网安备 11010802027423号
京公网安备 11010802027423号