Anaerobe ( IF 2.5 ) Pub Date : 2021-06-08 , DOI: 10.1016/j.anaerobe.2021.102403 Maryam Noori 1 , Zohreh Ghalavand 2 , Masoumeh Azimirad 3 , Abbas Yadegar 3 , Gita Eslami 2 , Marcela Krutova 4 , Marie Brajerova 4 , Mehdi Goudarzi 2 , Mohammad Reza Zali 5
|
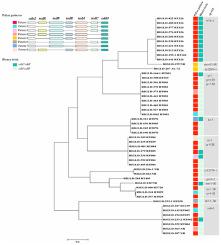
Clostridioides difficile is the most common causative agent of healthcare-associated diarrhea. C. difficile strains produce a crystalline surface layer protein (SlpA), encoded by the slpA gene. Previous studies have shown that SlpA varies among C. difficile strains. In this study, we used the SlpA sequence-based typing system (SlpAST) for the molecular genotyping of C. difficile clinical isolates identified in Iran; the PCR ribotypes (RTs) and toxin profiles of the isolates were also characterized. Forty-eight C. difficile isolates were obtained from diarrheal patients, and characterized by capillary electrophoresis (CE) PCR ribotyping and the detection of toxin genes. In addition, the genetic diversity of the slpA gene was investigated by Sanger sequencing. The most common RTs were RT126 (20.8%), followed by RT001 (12.5%) and RT084 (10.4%). The intact PaLoc arrangement representing cdu2+/tcdR+/tcdB+/tcdE+/tcdA+/tcdC+/cdd3+ profile was the predominant pattern and cdtA and cdtB genes were found in one-third of the isolates. Using the SlpA genotyping, 12 main genotypes and 16 subtypes were identified. The SlpA type 078–1 was the most prevalent genotype (20.8%), and identified within the isolates of RT126. The yok-1, gr-1, cr-1 and kr-3 genotypes were detected in 14.5%, 12.5%, 12.5% and 8.3% of isolates, respectively. Almost all the isolates with the same RT were clustered in similar SlpA sequence types. In comparison to PCR ribotyping, SlpAST, as a simple and highly reproducible sequenced-based technique, can discriminate well between C. difficile isolates. This typing method appears to be a valuable tool for the epidemiological study of C. difficile isolates worldwide.
中文翻译:

伊朗德黑兰艰难梭菌临床分离株表层蛋白A基因(slpA)的遗传多样性和系统发育分析
艰难梭菌是医疗保健相关性腹泻的最常见病原体。艰难梭菌菌株产生结晶表层蛋白 (SlpA),由slpA基因编码。先前的研究表明,SlpA 在艰难梭菌菌株之间有所不同。在本研究中,我们使用基于 SlpA 序列的分型系统 (SlpAST) 对在伊朗鉴定的艰难梭菌临床分离株进行分子基因分型;还表征了分离株的 PCR 核糖型 (RT) 和毒素谱。四十八C. difficile分离株来自腹泻患者,并通过毛细管电泳 (CE) PCR 核糖体分型和毒素基因检测表征。此外,通过Sanger测序研究了slpA基因的遗传多样性。最常见的 RT 是 RT126 (20.8%),其次是 RT001 (12.5%) 和 RT084 (10.4%)。代表cdu2 + / tcdR + / tcdB + / tcdE + / tcdA + / tcdC + / cdd3 +轮廓的完整 PaLoc 排列是主要模式,cdtA和cdtB在三分之一的分离株中发现了基因。使用 SlpA 基因分型,确定了 12 个主要基因型和 16 个亚型。SlpA 078-1 型是最普遍的基因型 (20.8%),并在 RT126 分离株中鉴定。yok-1、gr-1、cr-1 和 kr-3 基因型分别在 14.5%、12.5%、12.5% 和 8.3% 的分离株中检测到。几乎所有具有相同 RT 的分离株都聚集在相似的 SlpA 序列类型中。与 PCR 核糖体分型相比,SlpAST 作为一种简单且高度可重复的基于测序的技术,可以很好地区分艰难梭菌分离株。这种分型方法似乎是世界范围内艰难梭菌分离株流行病学研究的宝贵工具。
















































 京公网安备 11010802027423号
京公网安备 11010802027423号