Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dual Methylation-Sensitive Restriction Endonucleases Coupling with an RPA-Assisted CRISPR/Cas13a System (DESCS) for Highly Sensitive Analysis of DNA Methylation and Its Application for Point-of-Care Detection
ACS Sensors ( IF 8.2 ) Pub Date : 2021-05-21 , DOI: 10.1021/acssensors.1c00674 Xianfeng Wang 1 , Shiying Zhou 1 , Chengxiang Chu 1 , Mei Yang 1 , Danqun Huo 1, 2 , Changjun Hou 1, 2
ACS Sensors ( IF 8.2 ) Pub Date : 2021-05-21 , DOI: 10.1021/acssensors.1c00674 Xianfeng Wang 1 , Shiying Zhou 1 , Chengxiang Chu 1 , Mei Yang 1 , Danqun Huo 1, 2 , Changjun Hou 1, 2
Affiliation
|
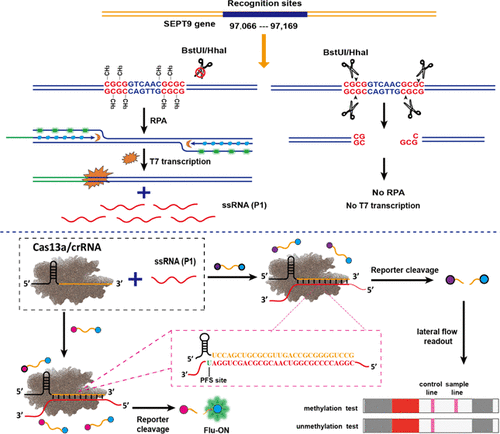
High-performance detection of DNA methylation possesses great significance for the diagnosis and therapy of cancer. Herein, for the first time, we present a digestion strategy based on dual methylation-sensitive restriction endonucleases coupling with a recombinase polymerase amplification (RPA)-assisted CRISPR/Cas13a system (DESCS) for accurate and sensitive determination of site-specific DNA methylation. This dual methylation-sensitive restriction endonuclease system selectively digests the unmethylated target but exhibits no response to methylated DNA. Therefore, the intact methylated DNA target triggers the RPA reaction for rapid signal amplification. In contrast, the digested unmethylated target initiates no RPA reaction. RPA products with a T7 promoter can execute the T7 transcription in the presence of T7 RNA polymerase to generate a large number of single-stranded RNA (ssRNA). This ssRNA can be recognized by CRISPR/Cas13a to induce the ssRNase activity of Cas13a, showing the indiscriminate cleavage of the collateral FQ reporter to release the fluorescence signal. With such a design, by combining the unique features of dual methylation-sensitive restriction endonucleases with RPA-assisted CRISPR/Cas13a, the DESCS system not only presents the rapid and powerful signal amplification for the determination of methylated DNA with ultrahigh sensitivity but also effectively eliminates the false positive influences from incomplete digestion of the unmethylated target. More importantly, 0.01% methylation level can be effectively distinguished with the existence of excess unmethylated DNA. In addition, the DESCS assay is integrated into the lateral flow biosensor (LFB) for the point-of-care determination of DNA methylation. In view of the superiorities in high sensitivity, outstanding selectivity, and ease of operation, the DESCS system will provide a reliable assay for site-specific analysis of methylation.
中文翻译:

双甲基化敏感限制性核酸内切酶与 RPA 辅助 CRISPR/Cas13a 系统 (DESCS) 偶联,用于 DNA 甲基化的高灵敏度分析及其在即时检测中的应用
DNA甲基化的高性能检测对于癌症的诊断和治疗具有重要意义。在此,我们首次提出了一种基于双甲基化敏感限制性内切核酸酶与重组酶聚合酶扩增 (RPA) 辅助 CRISPR/Cas13a 系统 (DESCS) 耦合的消化策略,用于准确和灵敏地确定位点特异性 DNA 甲基化。这种双重甲基化敏感性限制性内切核酸酶系统选择性地消化未甲基化的靶标,但对甲基化的 DNA 没有反应。因此,完整的甲基化 DNA 靶点会触发 RPA 反应以快速放大信号。相比之下,消化后的未甲基化目标不会引发 RPA 反应。带有T7启动子的RPA产物可以在T7 RNA聚合酶存在下执行T7转录,生成大量单链RNA(ssRNA)。这种 ssRNA 可以被 CRISPR/Cas13a 识别以诱导 Cas13a 的 ssRNase 活性,显示出对侧枝 FQ 报告基因的不加选择的切割以释放荧光信号。通过这样的设计,将双甲基化敏感限制性内切酶的独特特性与 RPA 辅助的 CRISPR/Cas13a 相结合,DESCS 系统不仅为甲基化 DNA 的测定提供了快速、强大的信号放大,而且具有超高灵敏度未甲基化目标的不完全消化造成的假阳性影响。更重要的是,0。01% 甲基化水平可以通过过量未甲基化 DNA 的存在有效区分。此外,DESCS 检测被集成到侧流生物传感器 (LFB) 中,用于即时检测 DNA 甲基化。鉴于在高灵敏度、出色的选择性和易于操作方面的优势,DESCS 系统将为甲基化的位点特异性分析提供可靠的检测方法。
更新日期:2021-06-25
中文翻译:

双甲基化敏感限制性核酸内切酶与 RPA 辅助 CRISPR/Cas13a 系统 (DESCS) 偶联,用于 DNA 甲基化的高灵敏度分析及其在即时检测中的应用
DNA甲基化的高性能检测对于癌症的诊断和治疗具有重要意义。在此,我们首次提出了一种基于双甲基化敏感限制性内切核酸酶与重组酶聚合酶扩增 (RPA) 辅助 CRISPR/Cas13a 系统 (DESCS) 耦合的消化策略,用于准确和灵敏地确定位点特异性 DNA 甲基化。这种双重甲基化敏感性限制性内切核酸酶系统选择性地消化未甲基化的靶标,但对甲基化的 DNA 没有反应。因此,完整的甲基化 DNA 靶点会触发 RPA 反应以快速放大信号。相比之下,消化后的未甲基化目标不会引发 RPA 反应。带有T7启动子的RPA产物可以在T7 RNA聚合酶存在下执行T7转录,生成大量单链RNA(ssRNA)。这种 ssRNA 可以被 CRISPR/Cas13a 识别以诱导 Cas13a 的 ssRNase 活性,显示出对侧枝 FQ 报告基因的不加选择的切割以释放荧光信号。通过这样的设计,将双甲基化敏感限制性内切酶的独特特性与 RPA 辅助的 CRISPR/Cas13a 相结合,DESCS 系统不仅为甲基化 DNA 的测定提供了快速、强大的信号放大,而且具有超高灵敏度未甲基化目标的不完全消化造成的假阳性影响。更重要的是,0。01% 甲基化水平可以通过过量未甲基化 DNA 的存在有效区分。此外,DESCS 检测被集成到侧流生物传感器 (LFB) 中,用于即时检测 DNA 甲基化。鉴于在高灵敏度、出色的选择性和易于操作方面的优势,DESCS 系统将为甲基化的位点特异性分析提供可靠的检测方法。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号