当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular approaches reveal weak sibship aggregation and a high dispersal propensity in a non-native fish parasite
Ecology and Evolution ( IF 2.3 ) Pub Date : 2021-05-06 , DOI: 10.1002/ece3.7415 Jérôme G Prunier 1 , Keoni Saint-Pé 1 , Simon Blanchet 1, 2 , Géraldine Loot 2 , Olivier Rey 3
Ecology and Evolution ( IF 2.3 ) Pub Date : 2021-05-06 , DOI: 10.1002/ece3.7415 Jérôme G Prunier 1 , Keoni Saint-Pé 1 , Simon Blanchet 1, 2 , Géraldine Loot 2 , Olivier Rey 3
Affiliation
|
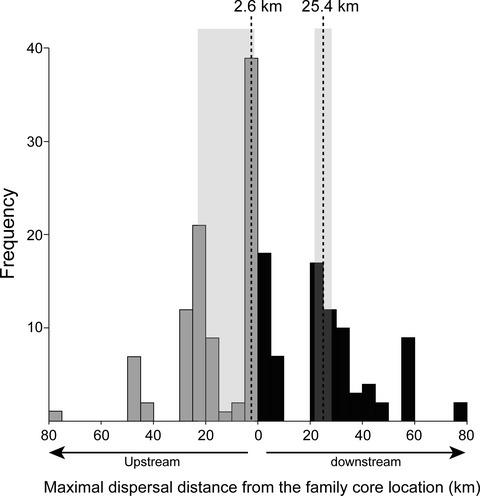
Inferring parameters related to the aggregation pattern of parasites and to their dispersal propensity are important for predicting their ecological consequences and evolutionary potential. Nonetheless, it is notoriously difficult to infer these parameters from wildlife parasites given the difficulty in tracking these organisms. Molecular-based inferences constitute a promising approach that has yet rarely been applied in the wild. Here, we combined several population genetic analyses including sibship reconstruction to document the genetic structure, patterns of sibship aggregation, and the dispersal dynamics of a non-native parasite of fish, the freshwater copepod ectoparasite Tracheliastes polycolpus. We collected parasites according to a hierarchical sampling design, with the sampling of all parasites from all host individuals captured in eight sites spread along an upstream–downstream river gradient. Individual multilocus genotypes were obtained from 14 microsatellite markers, and used to assign parasites to full-sib families and to investigate the genetic structure of T. polycolpus among both hosts and sampling sites. The distribution of full-sibs obtained among the sampling sites was used to estimate individual dispersal distances within families. Our results showed that T. polycolpus sibs tend to be aggregated within sites but not within host individuals. We detected important upstream-to-downstream dispersal events of T. polycolpus between sites (modal distance: 25.4 km; 95% CI [22.9, 27.7]), becoming scarcer as the geographic distance from their family core location increases. Such a dispersal pattern likely contributes to the strong isolation-by-distance observed at the river scale. We also detected some downstream-to-upstream dispersal events (modal distance: 2.6 km; 95% CI [2.2–23.3]) that likely result from movements of infected hosts. Within each site, the dispersal of free-living infective larvae among hosts likely contributes to increasing genetic diversity on hosts, possibly fostering the evolutionary potential of T. polycolpus.
中文翻译:

分子方法揭示非本地鱼类寄生虫的弱同胞聚集和高扩散倾向
推断与寄生虫聚集模式及其扩散倾向相关的参数对于预测其生态后果和进化潜力非常重要。尽管如此,鉴于追踪这些生物体的困难,从野生动物寄生虫中推断这些参数是出了名的困难。基于分子的推论是一种很有前途的方法,但目前很少在野外应用。在这里,我们结合了几种群体遗传分析,包括同胞关系重建,以记录遗传结构、同胞关系聚集模式以及非本地鱼类寄生虫(淡水桡足类外寄生虫Tracheliastes polycolpus)的扩散动态。我们根据分层抽样设计收集了寄生虫,从沿着上游-下游河流梯度分布的八个地点捕获的所有宿主个体中采集了所有寄生虫的样本。从 14 个微卫星标记获得个体多位点基因型,并用于将寄生虫分配到全同胞家族并研究T的遗传结构。宿主和采样点之间都有息肉。采样点中获得的全同胞分布用于估计家庭内个体的分散距离。我们的结果表明T .多沟氏菌同胞往往聚集在位点内,但不聚集在宿主个体内。我们检测到T的重要从上游到下游的扩散事件。地点之间的多沟(众数距离:25.4 km;95% CI [22.9, 27.7]),随着距家庭核心位置的地理距离增加而变得稀少。 这种分散模式可能导致了在河流尺度上观察到的强烈的距离隔离。我们还检测到一些下游到上游的传播事件(众数距离:2.6 公里;95% CI [2.2-23.3]),这可能是由受感染宿主的移动造成的。在每个地点内,自由生活的感染性幼虫在宿主之间的传播可能有助于增加宿主的遗传多样性,从而可能促进多沟毛虫的进化潜力。
更新日期:2021-06-17
中文翻译:

分子方法揭示非本地鱼类寄生虫的弱同胞聚集和高扩散倾向
推断与寄生虫聚集模式及其扩散倾向相关的参数对于预测其生态后果和进化潜力非常重要。尽管如此,鉴于追踪这些生物体的困难,从野生动物寄生虫中推断这些参数是出了名的困难。基于分子的推论是一种很有前途的方法,但目前很少在野外应用。在这里,我们结合了几种群体遗传分析,包括同胞关系重建,以记录遗传结构、同胞关系聚集模式以及非本地鱼类寄生虫(淡水桡足类外寄生虫Tracheliastes polycolpus)的扩散动态。我们根据分层抽样设计收集了寄生虫,从沿着上游-下游河流梯度分布的八个地点捕获的所有宿主个体中采集了所有寄生虫的样本。从 14 个微卫星标记获得个体多位点基因型,并用于将寄生虫分配到全同胞家族并研究T的遗传结构。宿主和采样点之间都有息肉。采样点中获得的全同胞分布用于估计家庭内个体的分散距离。我们的结果表明T .多沟氏菌同胞往往聚集在位点内,但不聚集在宿主个体内。我们检测到T的重要从上游到下游的扩散事件。地点之间的多沟(众数距离:25.4 km;95% CI [22.9, 27.7]),随着距家庭核心位置的地理距离增加而变得稀少。 这种分散模式可能导致了在河流尺度上观察到的强烈的距离隔离。我们还检测到一些下游到上游的传播事件(众数距离:2.6 公里;95% CI [2.2-23.3]),这可能是由受感染宿主的移动造成的。在每个地点内,自由生活的感染性幼虫在宿主之间的传播可能有助于增加宿主的遗传多样性,从而可能促进多沟毛虫的进化潜力。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号