当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5′-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid
Biochemistry ( IF 2.9 ) Pub Date : 2021-05-05 , DOI: 10.1021/acs.biochem.1c00106 Robert S Phillips 1, 2 , Samuel C-K Ting 2 , Kaitlin Anderson 3
Biochemistry ( IF 2.9 ) Pub Date : 2021-05-05 , DOI: 10.1021/acs.biochem.1c00106 Robert S Phillips 1, 2 , Samuel C-K Ting 2 , Kaitlin Anderson 3
Affiliation
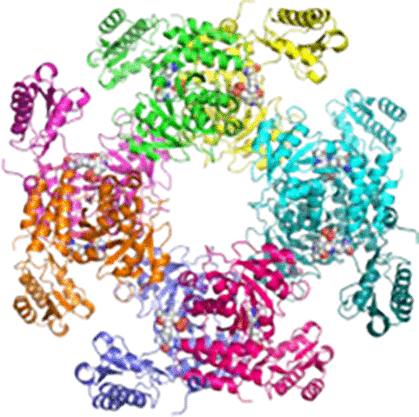
|
d-Glucosaminate-6-phosphate ammonia-lyase (DGL) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that produces 2-keto-3-deoxygluconate 6-phosphate (KDG-6-P) in the metabolism of d-glucosaminic acid by Salmonella enterica serovar typhimurium. We have determined the crystal structure of DGL by SAD phasing with selenomethionine to a resolution of 2.58 Å. The sequence has very low identity with most other members of the aminotransferase (AT) superfamily. The structure forms an octameric assembly as a tetramer of dimers that has not been observed previously in the AT superfamily. PLP is covalently bound as a Schiff base to Lys-213 in the catalytic dimer at the interface of two monomers. The structure lacks the conserved arginine that binds the α-carboxylate of the substrate in most members of the AT superfamily. However, there is a cluster of arginines in the small domain that likely serves as a binding site for the phosphate of the substrate. The deamination reaction performed in D2O gives a KDG-6-P product stereospecifically deuterated at C3; thus, the mechanism must involve an enamine intermediate that is protonated by the enzyme before product release. Nuclear magnetic resonance (NMR) analysis demonstrates that the deuterium is located in the pro-R position in the product, showing that the elimination of water takes place with inversion of configuration at C3, which is unprecedented for a PLP-dependent dehydratase/deaminase. On the basis of the crystal structure and the NMR data, a reaction mechanism for DGL is proposed.
中文翻译:

d-葡萄糖胺-6-磷酸氨裂解酶的结构和机制:吡哆醛 5'-磷酸依赖性酶的新型八聚体组装,以及 d-氨基酸消除反应中前所未有的立体化学反转
d -葡萄糖胺-6-磷酸氨裂解酶 (DGL) 是一种吡哆醛 5′-磷酸 (PLP) 依赖性酶,在d代谢中产生 2-酮-3-脱氧葡萄糖酸 6-磷酸 (KDG-6-P) -由鼠伤寒沙门氏菌产生的葡萄糖胺酸。我们通过硒代蛋氨酸 SAD 定相确定了 DGL 的晶体结构,分辨率为 2.58 Å。该序列与转氨酶 (AT) 超家族的大多数其他成员的同一性非常低。该结构形成八聚体组装体,作为二聚体的四聚体,此前在 AT 超家族中尚未观察到。 PLP 作为席夫碱与两个单体界面处催化二聚体中的 Lys-213 共价结合。在 AT 超家族的大多数成员中,该结构缺乏与底物 α-羧酸盐结合的保守精氨酸。然而,在小结构域中存在一簇精氨酸,可能充当底物磷酸盐的结合位点。在D 2 O中进行脱氨反应,得到C3立体定向氘化的KDG-6-P产物;因此,该机制必须涉及在产品释放之前被酶质子化的烯胺中间体。核磁共振(NMR)分析表明,氘位于产物中的pro - R位置,这表明水的消除是随着C3构型的反转而发生的,这对于PLP依赖性脱水酶/脱氨酶来说是前所未有的。根据晶体结构和 NMR 数据,提出了 DGL 的反应机理。
更新日期:2021-05-25
中文翻译:

d-葡萄糖胺-6-磷酸氨裂解酶的结构和机制:吡哆醛 5'-磷酸依赖性酶的新型八聚体组装,以及 d-氨基酸消除反应中前所未有的立体化学反转
d -葡萄糖胺-6-磷酸氨裂解酶 (DGL) 是一种吡哆醛 5′-磷酸 (PLP) 依赖性酶,在d代谢中产生 2-酮-3-脱氧葡萄糖酸 6-磷酸 (KDG-6-P) -由鼠伤寒沙门氏菌产生的葡萄糖胺酸。我们通过硒代蛋氨酸 SAD 定相确定了 DGL 的晶体结构,分辨率为 2.58 Å。该序列与转氨酶 (AT) 超家族的大多数其他成员的同一性非常低。该结构形成八聚体组装体,作为二聚体的四聚体,此前在 AT 超家族中尚未观察到。 PLP 作为席夫碱与两个单体界面处催化二聚体中的 Lys-213 共价结合。在 AT 超家族的大多数成员中,该结构缺乏与底物 α-羧酸盐结合的保守精氨酸。然而,在小结构域中存在一簇精氨酸,可能充当底物磷酸盐的结合位点。在D 2 O中进行脱氨反应,得到C3立体定向氘化的KDG-6-P产物;因此,该机制必须涉及在产品释放之前被酶质子化的烯胺中间体。核磁共振(NMR)分析表明,氘位于产物中的pro - R位置,这表明水的消除是随着C3构型的反转而发生的,这对于PLP依赖性脱水酶/脱氨酶来说是前所未有的。根据晶体结构和 NMR 数据,提出了 DGL 的反应机理。































 京公网安备 11010802027423号
京公网安备 11010802027423号