Anaerobe ( IF 2.5 ) Pub Date : 2021-02-20 , DOI: 10.1016/j.anaerobe.2021.102351
Leandro Gouveia Carneiro 1 , Tatiana Castro Abreu Pinto 2 , Hercules Moura 3 , John Barr 3 , Regina Maria Cavalcanti Pilotto Domingues 1 , Eliane de Oliveira Ferreira 1
|
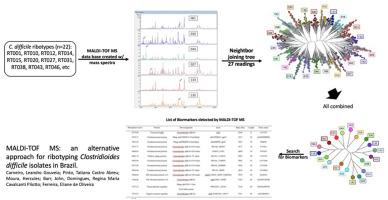
Clostridioides difficile is an important organism causing healthcare-associated infections. It has been documented that specific strains caused multiple outbreaks globally, and patients infected with those strains are more likely to develop severe C. difficile infection (CDI). With the appearance of a variant strain, BI/NAP1 ribotype 027, responsible for several outbreaks and high mortality rates worldwide, the epidemiology of the CDI changed drastically in the United States, Europe, and some Latin American countries. Although the epidemic strain 027 was not yet detected in Brazil, there are ribotypes exclusively found in the country, such as, 131, 132, 133, 135, 142 and 143, which are responsible for outbreaks in Brazilian hospitals and nursing homes. Although PCR-ribotyping is the most used method in epidemiology studies of C. difficile, it is not available in Brazil. This study aimed to develop and validate an in-house database for detecting C. difficile ribotypes, usually involved in CDI in Brazilian hospitals, by using MALDI-TOF MS. A database with 19 different ribotypes, 13 with worldwide circulation and 6 Brazilian-restricted, was created based on 27 spectra readings of each ribotype. After BioNumerics analysis, neighbor-joining trees revealed that spectra were distributed in clusters according to ribotypes, showing that MALDI-TOF MS could discriminate all 19 ribotypes. Moreover, each ribotype showed a different profile with 42 biomarkers detected in total. Based on their intensity and occurrence, 13 biomarkers were chosen to compose ribotype-specific profiles, and in silico analysis showed that most of these biomarkers were uncharacterized proteins or well-conserved peptides, such as ribosomal proteins. A double-blind assessment using the 13 biomarkers correctly assigned the ribotype in 73% of the spectra analyzed, with 94%–100% of correct hits for 027 and for Brazilian ribotypes. Although further analyses are required, our results show that MALDI-TOF MS might be a reliable, fast and feasible alternative for epidemiological surveillance of C. difficile in Brazil.
中文翻译:

MALDI-TOF MS:一种在巴西对艰难梭菌分离株进行核糖分型的替代方法
艰难梭菌是引起医疗保健相关感染的重要生物体。据记载,特定菌株在全球范围内引起了多次爆发,感染这些菌株的患者更有可能发展为严重的艰难梭菌感染(CDI)。随着变异菌株 BI/NAP1 核糖型 027 的出现,导致全球多次爆发和高死亡率,CDI 的流行病学在美国、欧洲和一些拉丁美洲国家发生了巨大变化。027型流行毒株虽然在巴西尚未发现,但存在巴西特有的核糖型,如131、132、133、135、142和143,是巴西医院和疗养院暴发的罪魁祸首。尽管 PCR 核糖分型是艰难梭菌流行病学研究中最常用的方法,但在巴西尚不可用。本研究旨在开发和验证用于检测艰难梭菌的内部数据库通过使用 MALDI-TOF MS,通常参与巴西医院的 CDI 的核糖型。基于每种核糖型的 27 个光谱读数,创建了一个包含 19 种不同核糖型的数据库,其中 13 种在世界范围内流通,6 种巴西限制使用。在 BioNumerics 分析后,邻接树显示光谱根据核糖体型分布在簇中,表明 MALDI-TOF MS 可以区分所有 19 种核糖体型。此外,每种核糖型显示出不同的特征,总共检测到 42 种生物标志物。根据它们的强度和发生率,选择了 13 种生物标志物来组成核型特异性图谱,并在计算机中分析表明,大多数这些生物标志物是未表征的蛋白质或保存完好的肽,例如核糖体蛋白。使用 13 种生物标志物进行的双盲评估在 73% 的分析光谱中正确分配了核型,027 和巴西核型的正确匹配率为 94%–100%。尽管需要进一步分析,但我们的结果表明,MALDI-TOF MS 可能是巴西艰难梭菌流行病学监测的可靠、快速和可行的替代方法。

































 京公网安备 11010802027423号
京公网安备 11010802027423号