Structure ( IF 4.4 ) Pub Date : 2021-02-03 , DOI: 10.1016/j.str.2021.01.005 Johnathan D Guest 1 , Thom Vreven 2 , Jing Zhou 3 , Iain Moal 4 , Jeliazko R Jeliazkov 5 , Jeffrey J Gray 6 , Zhiping Weng 2 , Brian G Pierce 1
|
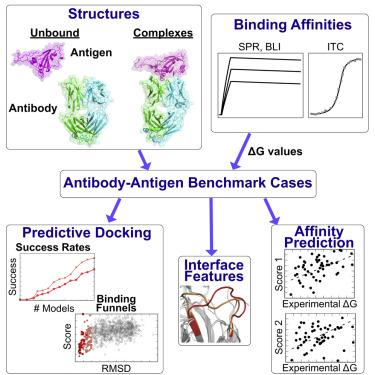
Accurate predictive modeling of antibody-antigen complex structures and structure-based antibody design remain major challenges in computational biology, with implications for biotherapeutics, immunity, and vaccines. Through a systematic search for high-resolution structures of antibody-antigen complexes and unbound antibody and antigen structures, in conjunction with identification of experimentally determined binding affinities, we have assembled a non-redundant set of test cases for antibody-antigen docking and affinity prediction. This benchmark more than doubles the number of antibody-antigen complexes and corresponding affinities available in our previous benchmarks, providing an unprecedented view of the determinants of antibody recognition and insights into molecular flexibility. Initial assessments of docking and affinity prediction tools highlight the challenges posed by this diverse set of cases, which includes camelid nanobodies, therapeutic monoclonal antibodies, and broadly neutralizing antibodies targeting viral glycoproteins. This dataset will enable development of advanced predictive modeling and design methods for this therapeutically relevant class of protein-protein interactions.
中文翻译:

抗体-抗原对接和亲和力预测的扩展基准揭示了对抗体识别决定因素的见解
抗体-抗原复杂结构的准确预测模型和基于结构的抗体设计仍然是计算生物学的主要挑战,对生物治疗、免疫和疫苗都有影响。通过系统搜索抗体-抗原复合物的高分辨率结构和未结合的抗体和抗原结构,结合实验确定的结合亲和力的鉴定,我们组装了一组用于抗体-抗原对接和亲和力预测的非冗余测试用例. 该基准使我们之前基准中可用的抗体-抗原复合物和相应亲和力的数量增加了一倍以上,为抗体识别的决定因素和分子灵活性提供了前所未有的视角。对接和亲和力预测工具的初步评估突出了这组不同案例带来的挑战,其中包括骆驼纳米抗体、治疗性单克隆抗体和针对病毒糖蛋白的广泛中和抗体。该数据集将有助于为这种治疗相关的蛋白质-蛋白质相互作用类别开发先进的预测建模和设计方法。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号