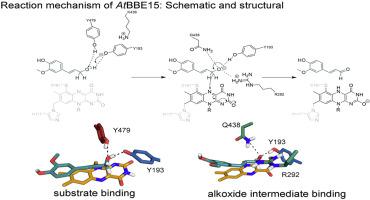
Monolignol氧化还原酶是黄连素桥酶样(BBE样)蛋白家族(pfam 08031)的成员,其将单木质酚氧化为相应的醛。它们是FAD依赖的酶,表现出对甲酚甲基羟化酶拓扑结构,也称为香草醛氧化酶拓扑结构。最近,我们曾报道从两个木质素单体氧化还原酶的结构和生化特性拟南芥,在BBE13和在BBE15。现在,我们对At进行了全面的定点诱变研究BBE15,以扩大我们对该酶类别的催化机理的了解。基于活性位点变体的动力学性质和分子动力学模拟,我们为单木香醇氧化还原酶家族提出了一种精炼的,结构指导的反应机理。在这里,我们建议通过烯丙醇的去质子化和随后的氢化物从醇盐的Cα原子转移到黄素上来促进该反应。我们描述了过多的氢键网络,使酶的催化机制。在该网络中,Tyr479和Tyr193协同作用为活性催化碱,以促进质子的提取。Lys436间接参与去质子化,因为该残基通过阳离子-π相互作用确定Tyr193的位置。该酶形成亲水腔,以容纳醇盐中间体并稳定从醇盐到醛的过渡态。通过分子动力学模拟,我们确定了醇和醇盐状态下底物的两种不同和不同的结合模式。醇与Tyr193和Tyr479相互作用,而Arg292,Gln438和Tyr193形成醇盐结合位点以容纳该中间体。活性位点变体活性的pH依赖性表明,醇盐结合位点的完整性对于p的微调也至关重要。我们已经确定了醇和醇盐状态的底物的两种不同和不同的结合模式。醇与Tyr193和Tyr479相互作用,而Arg292,Gln438和Tyr193形成醇盐结合位点以容纳该中间体。活性位点变体活性的pH依赖性表明,醇盐结合位点的完整性对于p的微调也至关重要。我们已经确定了醇和醇盐状态的底物的两种不同和不同的结合模式。醇与Tyr193和Tyr479相互作用,而Arg292,Gln438和Tyr193形成醇盐结合位点以容纳该中间体。活性位点变体活性的pH依赖性表明,醇盐结合位点的完整性对于p的微调也至关重要。Tyr193和Tyr479的K a。序列比对显示该机制的关键残基是高度保守的,表明我们提出的机制不仅与At BBE15有关,而且与大多数BBE-like蛋白有关。
 "点击查看英文标题和摘要"
"点击查看英文标题和摘要"
The catalytic machinery of the FAD-dependent AtBBE-like protein 15 for alcohol oxidation: Y193 and Y479 form a catalytic base, Q438 and R292 an alkoxide binding site
Monolignol oxidoreductases are members of the berberine bridge enzyme–like (BBE-like) protein family (pfam 08031) that oxidize monolignols to the corresponding aldehydes. They are FAD-dependent enzymes that exhibit the para-cresolmethylhydroxylase-topology, also known as vanillyl oxidase-topology. Recently, we have reported the structural and biochemical characterization of two monolignol oxidoreductases from Arabidopsis thaliana, AtBBE13 and AtBBE15. Now, we have conducted a comprehensive site directed mutagenesis study for AtBBE15, to expand our understanding of the catalytic mechanism of this enzyme class. Based on the kinetic properties of active site variants and molecular dynamics simulations, we propose a refined, structure-guided reaction mechanism for the family of monolignol oxidoreductases. Here, we propose that this reaction is facilitated stepwise by the deprotonation of the allylic alcohol and a subsequent hydride transfer from the Cα-atom of the alkoxide to the flavin. We describe an excessive hydrogen bond network that enables the catalytic mechanism of the enzyme. Within this network Tyr479 and Tyr193 act concertedly as active catalytic bases to facilitate the proton abstraction. Lys436 is indirectly involved in the deprotonation as this residue determines the position of Tyr193 via a cation-π interaction. The enzyme forms a hydrophilic cavity to accommodate the alkoxide intermediate and to stabilize the transition state from the alkoxide to the aldehyde. By means of molecular dynamics simulations, we have identified two different and distinct binding modes for the substrate in the alcohol and alkoxide state. The alcohol interacts with Tyr193 and Tyr479 while Arg292, Gln438 and Tyr193 form an alkoxide binding site to accommodate this intermediate. The pH-dependency of the activity of the active site variants revealed that the integrity of the alkoxide binding site is also crucial for the fine tuning of the pKa of Tyr193 and Tyr479. Sequence alignments showed that key residues for the mechanism are highly conserved, indicating that our proposed mechanism is not only relevant for AtBBE15 but for the majority of BBE-like proteins.
































 京公网安备 11010802027423号
京公网安备 11010802027423号