当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Systematic Quantification of Sequence and Structural Determinants Controlling mRNA stability in Bacterial Operons
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-01-19 , DOI: 10.1021/acssynbio.0c00471 Daniel P. Cetnar , Howard M. Salis
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-01-19 , DOI: 10.1021/acssynbio.0c00471 Daniel P. Cetnar , Howard M. Salis
|
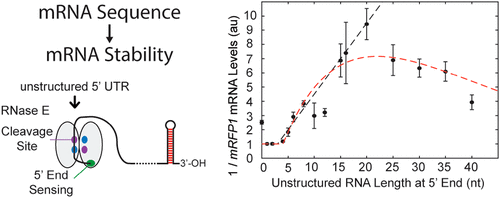
mRNA degradation is a central process that affects all gene expression levels, and yet, the determinants that control mRNA decay rates remain poorly characterized. Here, we applied a synthetic biology, learn-by-design approach to elucidate the sequence and structural determinants that control mRNA stability in bacterial operons. We designed, constructed, and characterized 82 operons in Escherichia coli, systematically varying RNase binding site characteristics, translation initiation rates, and transcriptional terminator efficiencies in the 5′ untranslated region (UTR), intergenic, and 3′ UTR regions, followed by measuring their mRNA levels using reverse transcription quantitative polymerase chain reaction (RT-qPCR) assays during exponential growth. We show that introducing long single-stranded RNA into 5′ UTRs reduced mRNA levels by up to 9.4-fold and that lowering translation rates reduced mRNA levels by up to 11.8-fold. We also found that RNase binding sites in intergenic regions had much lower effects on mRNA levels. Surprisingly, changing the transcriptional termination efficiency or introducing long single-stranded RNA into 3′ UTRs had no effect on upstream mRNA levels. From these measurements, we developed and validated biophysical models of ribosome protection and RNase activity with excellent quantitative agreement. We also formulated design rules to rationally control a mRNA’s stability, facilitating the automated design of engineered genetic systems with desired functionalities.
中文翻译:

控制细菌操纵子中 mRNA 稳定性的序列和结构决定因素的系统量化
mRNA 降解是影响所有基因表达水平的核心过程,然而,控制 mRNA 衰减率的决定因素仍然缺乏表征。在这里,我们应用合成生物学、设计学习方法来阐明控制细菌操纵子中 mRNA 稳定性的序列和结构决定因素。我们设计、构建并表征了大肠杆菌中的82 个操纵子,系统地改变 5' 非翻译区 (UTR)、基因间和 3' UTR 区域的 RNase 结合位点特征、翻译起始率和转录终止子效率,然后使用逆转录定量聚合酶链反应 (RT- qPCR) 检测在指数增长期间。我们表明,将长单链 RNA 引入 5' UTR 可将 mRNA 水平降低多达 9.4 倍,而降低翻译速率可将 mRNA 水平降低多达 11.8 倍。我们还发现基因间区域的 RNase 结合位点对 mRNA 水平的影响要低得多。令人惊讶的是,改变转录终止效率或将长单链 RNA 引入 3' UTR 对上游 mRNA 水平没有影响。从这些测量中,我们开发并验证了核糖体保护和 RNase 活性的生物物理模型,具有极好的定量一致性。我们还制定了设计规则来合理控制 mRNA 的稳定性,促进具有所需功能的工程遗传系统的自动化设计。
更新日期:2021-02-19
中文翻译:

控制细菌操纵子中 mRNA 稳定性的序列和结构决定因素的系统量化
mRNA 降解是影响所有基因表达水平的核心过程,然而,控制 mRNA 衰减率的决定因素仍然缺乏表征。在这里,我们应用合成生物学、设计学习方法来阐明控制细菌操纵子中 mRNA 稳定性的序列和结构决定因素。我们设计、构建并表征了大肠杆菌中的82 个操纵子,系统地改变 5' 非翻译区 (UTR)、基因间和 3' UTR 区域的 RNase 结合位点特征、翻译起始率和转录终止子效率,然后使用逆转录定量聚合酶链反应 (RT- qPCR) 检测在指数增长期间。我们表明,将长单链 RNA 引入 5' UTR 可将 mRNA 水平降低多达 9.4 倍,而降低翻译速率可将 mRNA 水平降低多达 11.8 倍。我们还发现基因间区域的 RNase 结合位点对 mRNA 水平的影响要低得多。令人惊讶的是,改变转录终止效率或将长单链 RNA 引入 3' UTR 对上游 mRNA 水平没有影响。从这些测量中,我们开发并验证了核糖体保护和 RNase 活性的生物物理模型,具有极好的定量一致性。我们还制定了设计规则来合理控制 mRNA 的稳定性,促进具有所需功能的工程遗传系统的自动化设计。















































 京公网安备 11010802027423号
京公网安备 11010802027423号