Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Multiplex Single-Molecule Kinetics of Nanopore-Coupled Polymerases
ACS Nano ( IF 15.8 ) Pub Date : 2020-12-28 , DOI: 10.1021/acsnano.0c05226 Mirkó Palla 1, 2 , Sukanya Punthambaker 1, 2 , Benjamin Stranges 1 , Frederic Vigneault 2 , Jeff Nivala 1 , Daniel Wiegand 2 , Aruna Ayer 3 , Timothy Craig 3 , Dmitriy Gremyachinskiy 3 , Helen Franklin 3 , Shaw Sun 3 , James Pollard 3 , Andrew Trans 3 , Cleoma Arnold 3 , Charles Schwab 3 , Colin Mcgaw 3 , Preethi Sarvabhowman 3 , Dhruti Dalal 3 , Eileen Thai 3 , Evan Amato 3 , Ilya Lederman 3 , Meng Taing 3 , Sara Kelley 3 , Adam Qwan 3 , Carl W Fuller 3, 4 , Stefan Roever 3 , George M Church 1, 2
ACS Nano ( IF 15.8 ) Pub Date : 2020-12-28 , DOI: 10.1021/acsnano.0c05226 Mirkó Palla 1, 2 , Sukanya Punthambaker 1, 2 , Benjamin Stranges 1 , Frederic Vigneault 2 , Jeff Nivala 1 , Daniel Wiegand 2 , Aruna Ayer 3 , Timothy Craig 3 , Dmitriy Gremyachinskiy 3 , Helen Franklin 3 , Shaw Sun 3 , James Pollard 3 , Andrew Trans 3 , Cleoma Arnold 3 , Charles Schwab 3 , Colin Mcgaw 3 , Preethi Sarvabhowman 3 , Dhruti Dalal 3 , Eileen Thai 3 , Evan Amato 3 , Ilya Lederman 3 , Meng Taing 3 , Sara Kelley 3 , Adam Qwan 3 , Carl W Fuller 3, 4 , Stefan Roever 3 , George M Church 1, 2
Affiliation
|
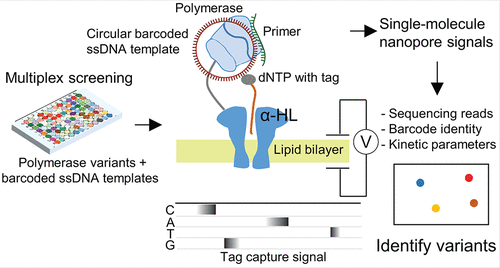
DNA polymerases have revolutionized the biotechnology field due to their ability to precisely replicate stored genetic information. Screening variants of these enzymes for specific properties gives the opportunity to identify polymerases with different features. We have previously developed a single-molecule DNA sequencing platform by coupling a DNA polymerase to an α-hemolysin pore on a nanopore array. Here, we use this approach to demonstrate a single-molecule method that enables rapid screening of polymerase variants in a multiplex manner. In this approach, barcoded DNA strands are complexed with polymerase variants and serve as templates for nanopore sequencing. Nanopore sequencing of the barcoded DNA reveals both the barcode identity and kinetic properties of the polymerase variant associated with the cognate barcode, allowing for multiplexed investigation of many polymerase variants in parallel on a single nanopore array. Further, we develop a robust classification algorithm that discriminates kinetic characteristics of the different polymerase mutants. As a proof of concept, we demonstrate the utility of our approach by screening a library of ∼100 polymerases to identify variants for potential applications of biotechnological interest. We anticipate our screening method to be broadly useful for applications that require polymerases with altered physical properties.
中文翻译:

纳米孔偶联聚合酶的多重单分子动力学
DNA 聚合酶由于能够精确复制存储的遗传信息而彻底改变了生物技术领域。筛选这些酶的特定特性的变体提供了识别具有不同特征的聚合酶的机会。我们之前通过将 DNA 聚合酶与纳米孔阵列上的 α-溶血素孔偶联,开发了单分子 DNA 测序平台。在这里,我们使用这种方法来演示一种单分子方法,该方法能够以多重方式快速筛选聚合酶变体。在这种方法中,带条形码的 DNA 链与聚合酶变体复合,并用作纳米孔测序的模板。条形码 DNA 的纳米孔测序揭示了与同源条形码相关的聚合酶变体的条形码身份和动力学特性,允许在单个纳米孔阵列上并行地对许多聚合酶变体进行多重研究。此外,我们开发了一种强大的分类算法,可以区分不同聚合酶突变体的动力学特征。作为概念证明,我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。
更新日期:2021-01-26
中文翻译:

纳米孔偶联聚合酶的多重单分子动力学
DNA 聚合酶由于能够精确复制存储的遗传信息而彻底改变了生物技术领域。筛选这些酶的特定特性的变体提供了识别具有不同特征的聚合酶的机会。我们之前通过将 DNA 聚合酶与纳米孔阵列上的 α-溶血素孔偶联,开发了单分子 DNA 测序平台。在这里,我们使用这种方法来演示一种单分子方法,该方法能够以多重方式快速筛选聚合酶变体。在这种方法中,带条形码的 DNA 链与聚合酶变体复合,并用作纳米孔测序的模板。条形码 DNA 的纳米孔测序揭示了与同源条形码相关的聚合酶变体的条形码身份和动力学特性,允许在单个纳米孔阵列上并行地对许多聚合酶变体进行多重研究。此外,我们开发了一种强大的分类算法,可以区分不同聚合酶突变体的动力学特征。作为概念证明,我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。我们通过筛选约 100 种聚合酶的文库来证明我们方法的实用性,以识别具有生物技术意义的潜在应用的变体。我们预计我们的筛选方法将广泛用于需要物理性质改变的聚合酶的应用。

































 京公网安备 11010802027423号
京公网安备 11010802027423号