Journal of Invertebrate Pathology ( IF 3.6 ) Pub Date : 2020-10-28 , DOI: 10.1016/j.jip.2020.107496 Aydin Yesilyurt 1 , Zihni Demirbag 2 , Monique M van Oers 3 , Remziye Nalcacioglu 2
|
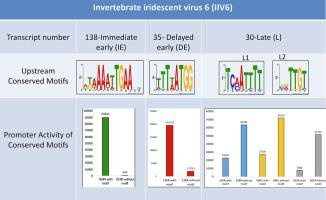
Invertebrate iridescent virus 6 (IIV6) is the type species of the Iridovirus genus in the Betairidovirinae subfamily of the Iridoviridae family. Transcription of the 215 predicted IIV6 genes is temporally regulated, dividing the genes into three kinetic classes: immediate-early (IE), delayed-early (DE), and late (L). So far, the transcriptional class has been determined for a selection of virion protein genes and only for three genes the potential promoter regions have been analyzed in detail. In this study, we investigated the transcriptional class of all IIV6 genes that had not been classified until now. RT-PCR analysis of total RNA isolated from virus-infected insect cells in the presence or absence of protein and DNA synthesis inhibitors, placed 113, 23 and 22 of the newly analyzed viral ORFs into the IE, DE and L gene classes, respectively. Afterwards, in silico analysis was performed to the upstream regions (200 bp) of all viral ORFs using the MEME Suite Software. The AA(A/T)(T/A)TG(A/G)A and (T/A/C)(T/G/C)T(T/A)ATGG motifs were identified in the upstream region of IE and DE genes, respectively. These motifs were validated by luciferase reporter assays as crucial sequences for promoter activity. For the L genes two conserved motifs were identified for all analyzed genes: (T/G)(C/T)(A/C)A(T/G/C)(T/C)T(T/C) and (C/G/T)(G/A/C)(T/A)(T/G) (G/T)(T/C). However, the presence of these two motifs did not influence promoter activity. Conversely, the presence of these two sequences upstream of the reporter decreased its expression. Single nucleotide mutations in the highly conserved nucleotides at the end of the second motif (TTGT) showed that this motif acted as a repressor sequence for late genes in the IIV6 genome. Next, upstream sequences of IIV6 L genes from which we removed this second motif in silico, were re-analyzed for the presence of potential conserved promoter sequences. Two additional motifs were identified in this way for L genes: (T/A)(A/T)(A/T/G)(A/T)(T/C)(A/G)(A/C)(A/C) and (C/G)(T/C)(T/A/C)C(A/T)(A/T)T(T/G) (T/G)(T/G/A). Independent mutations in either motif caused a severe decrease in luciferase expression. Information on temporal classes and upstream regulatory sequences will contribute to our understanding of the transcriptional mechanisms in IIV6.
中文翻译:

无脊椎动物虹彩病毒 6 (IIV6) 基因组中的保守基序调节病毒转录
无脊椎动物虹彩病毒 6 (IIV6) 是虹彩病毒科Betairidovirinae亚科中虹彩病毒属的模式种家庭。215 个预测的 IIV6 基因的转录受时间调控,将基因分为三个动力学类别:即刻早期 (IE)、延迟早期 (DE) 和晚期 (L)。到目前为止,已经为选择的病毒体蛋白基因确定了转录类别,并且仅对三个基因的潜在启动子区域进行了详细分析。在这项研究中,我们调查了迄今为止尚未分类的所有 IIV6 基因的转录类别。在存在或不存在蛋白质和 DNA 合成抑制剂的情况下,对从病毒感染的昆虫细胞中分离的总 RNA 进行 RT-PCR 分析,将新分析的病毒 ORF 中的 113、23 和 22 分别放入 IE、DE 和 L 基因类别。之后,在硅使用 MEME Suite 软件对所有病毒 ORF 的上游区域 (200 bp) 进行分析。在 IE 的上游区域鉴定了 AA(A/T)(T/A)TG(A/G)A 和 (T/A/C)(T/G/C)T(T/A)ATGG 基序和 DE 基因。这些基序通过萤光素酶报告基因检测验证为启动子活性的关键序列。对于 L 基因,为所有分析的基因鉴定了两个保守基序:(T/G)(C/T)(A/C)A(T/G/C)(T/C)T(T/C) 和( C/G/T)(G/A/C)(T/A)(T/G) (G/T)(T/C)。然而,这两个基序的存在不影响启动子活性。相反,报告基因上游的这两个序列的存在降低了其表达。第二个基序 (TTGT) 末端高度保守的核苷酸中的单核苷酸突变表明该基序充当 IIV6 基因组中晚期基因的阻遏序列。接下来,我们从中移除了第二个基序的 IIV6 L 基因的上游序列在计算机上,重新分析了潜在保守启动子序列的存在。以这种方式为 L 基因鉴定了两个额外的基序:(T/A)(A/T)(A/T/G)(A/T)(T/C)(A/G)(A/C)( A/C) 和 (C/G)(T/C)(T/A/C)C(A/T)(A/T)T(T/G) (T/G)(T/G/A )。任一基序中的独立突变都导致荧光素酶表达严重下降。有关时间类别和上游调控序列的信息将有助于我们了解 IIV6 中的转录机制。

































 京公网安备 11010802027423号
京公网安备 11010802027423号