当前位置:
X-MOL 学术
›
Cancer Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genome-wide DNA methylation analysis by MethylRad and the transcriptome profiles reveal the potential cancer-related lncRNAs in colon cancer.
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-09-01 , DOI: 10.1002/cam4.3412 Guixi Zheng 1 , Yuzhi Zhang 2 , Hongchun Wang 1 , E Ding 1 , Ailin Qu 1 , Peng Su 3 , Yongmei Yang 1 , Mingjin Zou 1 , Yi Zhang 1
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-09-01 , DOI: 10.1002/cam4.3412 Guixi Zheng 1 , Yuzhi Zhang 2 , Hongchun Wang 1 , E Ding 1 , Ailin Qu 1 , Peng Su 3 , Yongmei Yang 1 , Mingjin Zou 1 , Yi Zhang 1
Affiliation
|
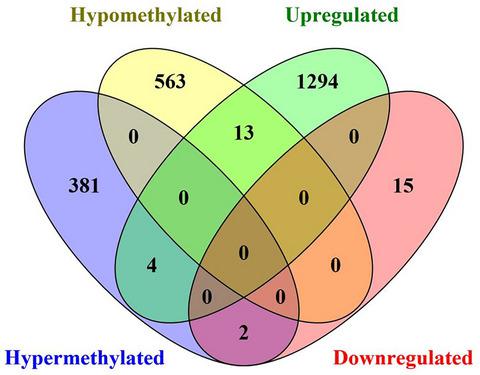
Colon cancer (CC) is characterized by global aberrant DNA methylation that may affect gene expression and genomic stability. A series of studies have demonstrated that DNA methylation could regulate the expressions of not only protein‐coding genes but also ncRNAs. However, the regulatory role of lncRNA genes methylaton in CC remains largely unknown. In the present study, we systemically characterize the profile of DNA methylation, especially the aberrant methylation of lncRNAs genes using MethylRAD technology. A total of 132 999 CCGG/8487 CCWGG sites were identified as differentially methylated sites (DMSs), which were mainly located on the introns and intergenic elements. Moreover, 1,359 CCGG/1,052 CCWGG differentially methylated genes (DMGs) were screened. Our results demonstrated that aberrant methylation of lncRNA genes occurred most frequently, accounting for 37.5% and 44.3% in CCGG and CCWGG DMGs respectively. In addition, 963 lncRNA DMGs were co‐analyzed with 1328 differentially expressed lncRNAs which were identified from TCGA database. We found that 15 lncRNAs might be CC‐related lncRNAs. ZNF667‐AS1 and MAFA‐AS1 were down‐regulated in CC, which might be silenced by hypermethylation. Besides, 13 lncRNAs were hypomethylated and up‐regulated in CC. Moreover, our results validated the expression and methylation level of CC‐related lncRNAs by RT‐qPCR and pyrosequencing assay. In conclusion, we performed a genome‐wide DNA methylation analysis by MethylRAD to acquire both CCGG and CCWGG DMSs and DMGs in CC. The results screened lncRNA DMSs as potential biomarkers and identified 15 lncRNAs as CC‐related lncRNAs. This study provided novel therapy targets and valuable insights into molecular mechanism in tumorigenesis and development of CC.
中文翻译:

通过MethylRad进行的全基因组DNA甲基化分析和转录组图谱揭示了结肠癌中潜在的与癌症相关的lncRNA。
结肠癌(CC)的特征在于整体异常的DNA甲基化,这可能会影响基因表达和基因组稳定性。一系列研究表明,DNA甲基化不仅可以调节蛋白质编码基因的表达,还可以调节ncRNA的表达。然而,lncRNA基因甲基酮在CC中的调控作用仍然未知。在本研究中,我们使用MethylRAD技术系统地表征了DNA甲基化的特征,尤其是lncRNAs基因的异常甲基化。总共132 999 CCGG / 8487 CCWGG位点被鉴定为差异甲基化位点(DMS),主要位于内含子和基因间元件上。此外,筛选了1,359个CCGG / 1,052个CCWGG差异甲基化基因(DMG)。我们的结果表明,lncRNA基因异常甲基化发生最频繁,分别占CCGG和CCWGG DMG的37.5%和44.3%。此外,还对963个lncRNA DMG与1328个差异表达的lncRNA进行了共同分析,这些tncRNA是从TCGA数据库中鉴定的。我们发现15个lncRNA可能是CC相关的lncRNA。ZNF667-AS1和MAFA-AS1在CC中被下调,而高甲基化可能会使其沉默。此外,CC中有13个lncRNA被低甲基化和上调。此外,我们的结果通过RT-qPCR和焦磷酸测序法验证了CC相关lncRNA的表达和甲基化水平。最后,我们通过MethylRAD进行了全基因组DNA甲基化分析,以获取CC中的CCGG和CCWGG DMS和DMG。结果筛选出了lncRNA DMS作为潜在的生物标志物,并鉴定出15个lncRNA为CC相关的lncRNA。这项研究提供了新颖的治疗靶点和对CC发生和发展的分子机制的宝贵见解。
更新日期:2020-10-19
中文翻译:

通过MethylRad进行的全基因组DNA甲基化分析和转录组图谱揭示了结肠癌中潜在的与癌症相关的lncRNA。
结肠癌(CC)的特征在于整体异常的DNA甲基化,这可能会影响基因表达和基因组稳定性。一系列研究表明,DNA甲基化不仅可以调节蛋白质编码基因的表达,还可以调节ncRNA的表达。然而,lncRNA基因甲基酮在CC中的调控作用仍然未知。在本研究中,我们使用MethylRAD技术系统地表征了DNA甲基化的特征,尤其是lncRNAs基因的异常甲基化。总共132 999 CCGG / 8487 CCWGG位点被鉴定为差异甲基化位点(DMS),主要位于内含子和基因间元件上。此外,筛选了1,359个CCGG / 1,052个CCWGG差异甲基化基因(DMG)。我们的结果表明,lncRNA基因异常甲基化发生最频繁,分别占CCGG和CCWGG DMG的37.5%和44.3%。此外,还对963个lncRNA DMG与1328个差异表达的lncRNA进行了共同分析,这些tncRNA是从TCGA数据库中鉴定的。我们发现15个lncRNA可能是CC相关的lncRNA。ZNF667-AS1和MAFA-AS1在CC中被下调,而高甲基化可能会使其沉默。此外,CC中有13个lncRNA被低甲基化和上调。此外,我们的结果通过RT-qPCR和焦磷酸测序法验证了CC相关lncRNA的表达和甲基化水平。最后,我们通过MethylRAD进行了全基因组DNA甲基化分析,以获取CC中的CCGG和CCWGG DMS和DMG。结果筛选出了lncRNA DMS作为潜在的生物标志物,并鉴定出15个lncRNA为CC相关的lncRNA。这项研究提供了新颖的治疗靶点和对CC发生和发展的分子机制的宝贵见解。















































 京公网安备 11010802027423号
京公网安备 11010802027423号