Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis.
The Plant Journal ( IF 6.2 ) Pub Date : 2020-06-10 , DOI: 10.1111/tpj.14874
Qundan Lv 1 , Jie Qiu 2 , Jie Liu 2 , Zheng Li 3 , Wenting Zhang 2 , Qin Wang 2 , Jie Fang 1 , Junjie Pan 1 , Zhengdao Chen 1 , Wenliang Cheng 1 , Michael S Barker 3 , Xuehui Huang 2 , Xin Wei 2 , Kejun Cheng 1
The Plant Journal ( IF 6.2 ) Pub Date : 2020-06-10 , DOI: 10.1111/tpj.14874
Qundan Lv 1 , Jie Qiu 2 , Jie Liu 2 , Zheng Li 3 , Wenting Zhang 2 , Qin Wang 2 , Jie Fang 1 , Junjie Pan 1 , Zhengdao Chen 1 , Wenliang Cheng 1 , Michael S Barker 3 , Xuehui Huang 2 , Xin Wei 2 , Kejun Cheng 1
Affiliation
|
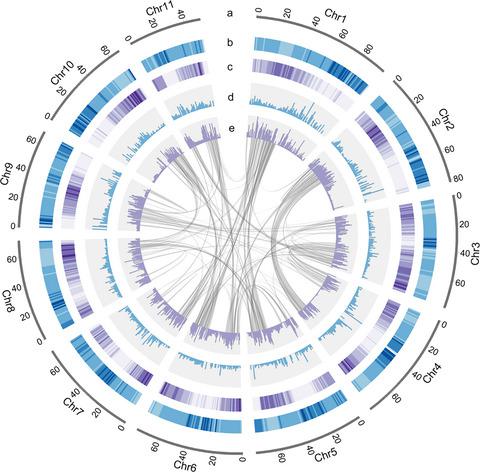
Chimonanthus salicifolius, a member of the Calycanthaceae of magnoliids, is one of the most famous medicinal plants in Eastern China. Here, we report a chromosome‐level genome assembly of C. salicifolius, comprising 820.1 Mb of genomic sequence with a contig N50 of 2.3 Mb and containing 36 651 annotated protein‐coding genes. Phylogenetic analyses revealed that magnoliids were sister to the eudicots. Two rounds of ancient whole‐genome duplication were inferred in the C. salicifolious genome. One is shared by Calycanthaceae after its divergence with Lauraceae, and the other is in the ancestry of Magnoliales and Laurales. Notably, long genes with > 20 kb in length were much more prevalent in the magnoliid genomes compared with other angiosperms, which could be caused by the length expansion of introns inserted by transposon elements. Homologous genes within the flavonoid pathway for C. salicifolius were identified, and correlation of the gene expression and the contents of flavonoid metabolites revealed potential critical genes involved in flavonoids biosynthesis. This study not only provides an additional whole‐genome sequence from the magnoliids, but also opens the door to functional genomic research and molecular breeding of C. salicifolius.
更新日期:2020-06-10

































 京公网安备 11010802027423号
京公网安备 11010802027423号