Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Programming Biomimetically Confined Aptamers with DNA Frameworks.
ACS Nano ( IF 15.8 ) Pub Date : 2020-06-02 , DOI: 10.1021/acsnano.0c03362 Xiuhai Mao 1 , Mengmeng Liu 2 , Lei Yan 1 , Mengying Deng 3, 4 , Fan Li 1 , Min Li 1 , Fei Wang 5, 6 , Jiang Li 3, 4 , Lihua Wang 3, 4 , Yang Tian 2 , Chunhai Fan 1, 6 , Xiaolei Zuo 1, 6
ACS Nano ( IF 15.8 ) Pub Date : 2020-06-02 , DOI: 10.1021/acsnano.0c03362 Xiuhai Mao 1 , Mengmeng Liu 2 , Lei Yan 1 , Mengying Deng 3, 4 , Fan Li 1 , Min Li 1 , Fei Wang 5, 6 , Jiang Li 3, 4 , Lihua Wang 3, 4 , Yang Tian 2 , Chunhai Fan 1, 6 , Xiaolei Zuo 1, 6
Affiliation
|
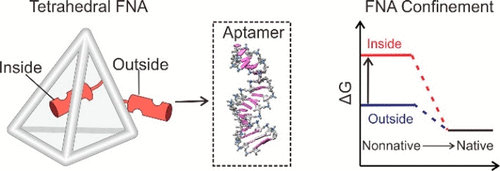
Active sites of proteins are generally encapsulated within three-dimensional peptide scaffolds that provide the molecular-scale confinement microenvironment. Nevertheless, the ability to tune thermodynamic stability in biomimetic molecular confinement relies on the macromolecular crowding effect of lack of stoichiometry and reconfigurability. Here, we report a framework nucleic acid (FNA)-based strategy to increase thermodynamic stability of aptamers. We demonstrate that the molecular-scale confinement increases the thermodynamic stability of aptamers via facilitated folding kinetics, which is confirmed by the single-molecule FRET (smFRET). Unfavorable conformations of aptamers are restricted as revealed by the Monte Carlo simulation. The binding affinity of the DNA framework-confined aptamer is improved by ∼3-fold. With a similar strategy we improve the catalytic activity of hemin-binding aptamer. Our approach thus shows high potential for designing protein-mimicking DNA nanostructures with enhanced binding affinity and catalytic activity for biosensing and biomedical engineering.
中文翻译:

用DNA框架编程仿生限制的适体。
蛋白质的活性位点通常被封装在三维分子支架中,该分子支架提供了分子级限制的微环境。然而,在仿生分子约束下调节热力学稳定性的能力取决于缺乏化学计量和可重构性的大分子拥挤效应。在这里,我们报告基于框架核酸(FNA)的策略,以增加适体的热力学稳定性。我们表明,分子尺度限制增加适体的热力学稳定性通过单分子FRET(smFRET)证实了其促进的折叠动力学。如蒙特卡洛模拟所揭示的,适体的不利构象受到限制。DNA框架限定的适体的结合亲和力提高了约3倍。通过类似的策略,我们提高了血红素结合适体的催化活性。因此,我们的方法显示出在设计具有模拟结合力的蛋白质纳米DNA纳米结构方面具有很高的潜力,这种纳米结构具有增强的结合亲和力和催化活性,可用于生物传感和生物医学工程。
更新日期:2020-07-28
中文翻译:

用DNA框架编程仿生限制的适体。
蛋白质的活性位点通常被封装在三维分子支架中,该分子支架提供了分子级限制的微环境。然而,在仿生分子约束下调节热力学稳定性的能力取决于缺乏化学计量和可重构性的大分子拥挤效应。在这里,我们报告基于框架核酸(FNA)的策略,以增加适体的热力学稳定性。我们表明,分子尺度限制增加适体的热力学稳定性通过单分子FRET(smFRET)证实了其促进的折叠动力学。如蒙特卡洛模拟所揭示的,适体的不利构象受到限制。DNA框架限定的适体的结合亲和力提高了约3倍。通过类似的策略,我们提高了血红素结合适体的催化活性。因此,我们的方法显示出在设计具有模拟结合力的蛋白质纳米DNA纳米结构方面具有很高的潜力,这种纳米结构具有增强的结合亲和力和催化活性,可用于生物传感和生物医学工程。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号