当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Improved Discrimination of Asymmetric and Symmetric Arginine Dimethylation by Optimization of the Normalized Collision Energy in Liquid Chromatography-Mass Spectrometry Proteomics.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-05-19 , DOI: 10.1021/acs.jproteome.0c00116
Nicolas G Hartel 1 , Christopher Z Liu 1 , Nicholas A Graham 1, 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-05-19 , DOI: 10.1021/acs.jproteome.0c00116
Nicolas G Hartel 1 , Christopher Z Liu 1 , Nicholas A Graham 1, 2
Affiliation
|
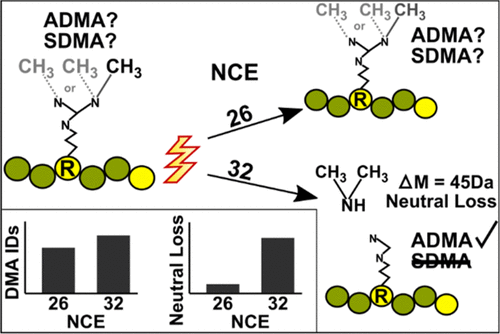
Protein arginine methylation regulates diverse biological processes including signaling, metabolism, splicing, and transcription. Despite its important biological roles, arginine dimethylation remains an understudied post-translational modification. Partly, this is because the two forms of arginine dimethylation, asymmetric dimethylarginine (ADMA) and symmetric dimethylarginine (SDMA), are isobaric and therefore indistinguishable by traditional mass spectrometry techniques. Thus, there exists a need for methods that can differentiate these two modifications. Recently, it has been shown that the ADMA and SDMA can be distinguished by the characteristic neutral loss (NL) of dimethylamine and methylamine, respectively. However, the utility of this method is limited because the vast majority of dimethylarginine peptides do not generate measurable NL ions. Here, we report that increasing the normalized collision energy (NCE) in a higher-energy collisional dissociation cell increases the generation of the characteristic NLs that distinguish ADMA and SDMA. By analyzing both synthetic and endogenous methyl-peptides, we identify an optimal NCE value that maximizes NL generation and simultaneously improves methyl-peptide identification. Using two orthogonal methyl-peptide enrichment strategies, high pH strong cation-exchange and immunoaffinity purification, we demonstrate that the optimal NCE improves NL-based ADMA and SDMA annotation and dimethyl-peptide identifications by 125% and 17%, respectively, compared to the standard NCE. This simple parameter change will greatly facilitate the identification and annotation of ADMA and SDMA in mass spectrometry-based methyl-proteomics to improve our understanding of how these modifications differentially regulate protein function. All raw data have been deposited in the PRIDE database with accession number PXD017193.
中文翻译:

通过优化液相色谱-质谱法蛋白质组学中的归一化碰撞能,改进了对不对称和对称精氨酸二甲基化的区分。
蛋白质精氨酸甲基化调节多种生物过程,包括信号传导,代谢,剪接和转录。尽管其重要的生物学作用,精氨酸二甲基化仍然是一个尚未被研究的翻译后修饰。部分原因是因为精氨酸二甲基化的两种形式,即不对称二甲基精氨酸(ADMA)和对称二甲基精氨酸(SDMA)等压,因此无法通过传统质谱技术区分。因此,需要可以区分这两种修改的方法。最近,已经表明,ADMA和SDMA可以分别通过二甲胺和甲胺的特征性中性损失(NL)来区分。然而,该方法的实用性受到限制,因为绝大多数二甲基精氨酸肽无法产生可测量的NL离子。在这里,我们报告说,在高能碰撞解离单元中增加归一化碰撞能量(NCE)会增加区分ADMA和SDMA的特征NL的生成。通过分析合成和内源性甲基肽,我们确定了一个最佳的NCE值,该值可最大化NL生成并同时提高甲基肽的识别率。使用两种正交的甲基肽富集策略,高pH值的强阳离子交换和免疫亲和纯化,我们证明了最佳NCE相比于NL,将基于NL的ADMA和SDMA注释以及二甲基肽鉴定分别提高了125%和17%。标准NCE。这种简单的参数更改将极大地促进基于质谱的甲基蛋白质组学中ADMA和SDMA的识别和注释,从而增进我们对这些修饰如何差异调节蛋白质功能的理解。所有原始数据都已保存在PRIDE数据库中,登录号为PXD017193。
更新日期:2020-05-19
中文翻译:

通过优化液相色谱-质谱法蛋白质组学中的归一化碰撞能,改进了对不对称和对称精氨酸二甲基化的区分。
蛋白质精氨酸甲基化调节多种生物过程,包括信号传导,代谢,剪接和转录。尽管其重要的生物学作用,精氨酸二甲基化仍然是一个尚未被研究的翻译后修饰。部分原因是因为精氨酸二甲基化的两种形式,即不对称二甲基精氨酸(ADMA)和对称二甲基精氨酸(SDMA)等压,因此无法通过传统质谱技术区分。因此,需要可以区分这两种修改的方法。最近,已经表明,ADMA和SDMA可以分别通过二甲胺和甲胺的特征性中性损失(NL)来区分。然而,该方法的实用性受到限制,因为绝大多数二甲基精氨酸肽无法产生可测量的NL离子。在这里,我们报告说,在高能碰撞解离单元中增加归一化碰撞能量(NCE)会增加区分ADMA和SDMA的特征NL的生成。通过分析合成和内源性甲基肽,我们确定了一个最佳的NCE值,该值可最大化NL生成并同时提高甲基肽的识别率。使用两种正交的甲基肽富集策略,高pH值的强阳离子交换和免疫亲和纯化,我们证明了最佳NCE相比于NL,将基于NL的ADMA和SDMA注释以及二甲基肽鉴定分别提高了125%和17%。标准NCE。这种简单的参数更改将极大地促进基于质谱的甲基蛋白质组学中ADMA和SDMA的识别和注释,从而增进我们对这些修饰如何差异调节蛋白质功能的理解。所有原始数据都已保存在PRIDE数据库中,登录号为PXD017193。































 京公网安备 11010802027423号
京公网安备 11010802027423号