当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Quantitative cooperative binding model for intrinsically disordered proteins interacting with nanomaterials
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2020-05-19 , DOI: 10.1021/jacs.0c01885 Da-Wei Li 1 , Mouzhe Xie 2 , Rafael Brüschweiler 1, 2, 3
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2020-05-19 , DOI: 10.1021/jacs.0c01885 Da-Wei Li 1 , Mouzhe Xie 2 , Rafael Brüschweiler 1, 2, 3
Affiliation
|
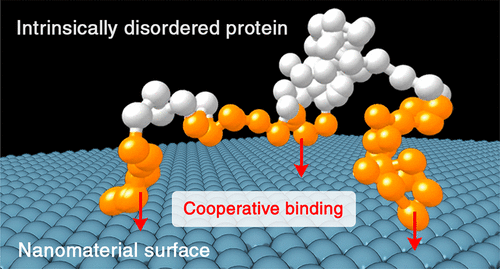
Intrinsically disordered proteins (IDPs) can display a broad spectrum of binding modes and highly variable binding affinities when interacting with both biological and non-biological materials. A quantitative model of such behavior is important for the better understanding of the function of IDPs when encountering inorganic nanomaterials with the potential to control their behavior in vivo and in vitro. Depending on their amino-acid composition and chain length, binding properties can vary strongly between different IDPs. Moreover, due to differences in the physical chemical properties of clusters of amino-acid residues along the IDP primary sequence, individual residues can adopt a wide range of bound state populations. Quantitative experimental binding affinities with synthetic silica nanoparticles (SNPs) at residue-level resolution, which were obtained for a set of IDPs by solution NMR relaxation experiments, are explained here by a first-principle analytical statistical mechanical model termed SILC. SILC quantitatively predicts residue-specific binding affinities to nanoparticles and it expresses binding cooperativity as the cumulative result of pairwise residue effects. The model, which was parametrized for anionic SNPs and applied to experimental data of four IDP systems with distinctive binding behavior, successfully predicts differences in overall binding affinities, fine details of IDP-SNP affinity profiles, and site-directed mutagenesis effects with a spatial resolution at the individual residue level. The SILC model provides an analytical description of such types of fuzzy IDP-SNP complexes and may help advance understanding nanotoxicity and in-vivo targeting of IDPs by specifically designed nanomaterials.
中文翻译:

固有无序蛋白质与纳米材料相互作用的定量协同结合模型
当与生物和非生物材料相互作用时,固有无序蛋白 (IDP) 可以显示出广泛的结合模式和高度可变的结合亲和力。当遇到具有控制其体内和体外行为的潜力的无机纳米材料时,这种行为的定量模型对于更好地理解 IDP 的功能很重要。根据它们的氨基酸组成和链长,不同 IDP 之间的结合特性可能会有很大差异。此外,由于沿 IDP 一级序列的氨基酸残基簇的物理化学性质的差异,单个残基可以采用广泛的结合状态群体。在残留水平分辨率下与合成二氧化硅纳米粒子 (SNP) 的定量实验结合亲和力,这是通过溶液 NMR 弛豫实验获得的一组 IDP,这里通过称为 SILC 的第一原理分析统计力学模型进行解释。SILC 定量预测与纳米颗粒的特定残基结合亲和力,并将结合协同性表示为成对残基效应的累积结果。该模型针对阴离子 SNP 参数化并应用于具有独特结合行为的四种 IDP 系统的实验数据,成功预测了整体结合亲和力的差异、IDP-SNP 亲和力谱的细节以及具有空间分辨率的定点诱变效应在个别残留水平。
更新日期:2020-05-19
中文翻译:

固有无序蛋白质与纳米材料相互作用的定量协同结合模型
当与生物和非生物材料相互作用时,固有无序蛋白 (IDP) 可以显示出广泛的结合模式和高度可变的结合亲和力。当遇到具有控制其体内和体外行为的潜力的无机纳米材料时,这种行为的定量模型对于更好地理解 IDP 的功能很重要。根据它们的氨基酸组成和链长,不同 IDP 之间的结合特性可能会有很大差异。此外,由于沿 IDP 一级序列的氨基酸残基簇的物理化学性质的差异,单个残基可以采用广泛的结合状态群体。在残留水平分辨率下与合成二氧化硅纳米粒子 (SNP) 的定量实验结合亲和力,这是通过溶液 NMR 弛豫实验获得的一组 IDP,这里通过称为 SILC 的第一原理分析统计力学模型进行解释。SILC 定量预测与纳米颗粒的特定残基结合亲和力,并将结合协同性表示为成对残基效应的累积结果。该模型针对阴离子 SNP 参数化并应用于具有独特结合行为的四种 IDP 系统的实验数据,成功预测了整体结合亲和力的差异、IDP-SNP 亲和力谱的细节以及具有空间分辨率的定点诱变效应在个别残留水平。

































 京公网安备 11010802027423号
京公网安备 11010802027423号