当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Brain tumor classification of virtual NMR voxels based on realistic blood vessel-induced spin dephasing using support vector machines
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-04-14 , DOI: 10.1002/nbm.4307 Artur Hahn 1, 2 , Julia Bode 3 , Sarah Schuhegger 1, 2 , Thomas Krüwel 3 , Volker J F Sturm 1, 4 , Ke Zhang 1, 4 , Johann M E Jende 1 , Björn Tews 3 , Sabine Heiland 1 , Martin Bendszus 1 , Michael O Breckwoldt 1, 5 , Christian H Ziener 1, 4 , Felix T Kurz 1, 4
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-04-14 , DOI: 10.1002/nbm.4307 Artur Hahn 1, 2 , Julia Bode 3 , Sarah Schuhegger 1, 2 , Thomas Krüwel 3 , Volker J F Sturm 1, 4 , Ke Zhang 1, 4 , Johann M E Jende 1 , Björn Tews 3 , Sabine Heiland 1 , Martin Bendszus 1 , Michael O Breckwoldt 1, 5 , Christian H Ziener 1, 4 , Felix T Kurz 1, 4
Affiliation
|
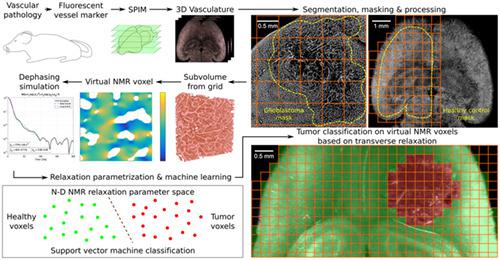
Remodeling of tissue microvasculature commonly promotes neoplastic growth; however, there is no imaging modality in oncology yet that noninvasively quantifies microvascular changes in clinical routine. Although blood capillaries cannot be resolved in typical magnetic resonance imaging (MRI) measurements, their geometry and distribution influence the integral nuclear magnetic resonance (NMR) signal from each macroscopic MRI voxel. We have numerically simulated the expected transverse relaxation in NMR voxels with different dimensions based on the realistic microvasculature in healthy and tumor-bearing mouse brains (U87 and GL261 glioblastoma). The 3D capillary structure in entire, undissected brains was acquired using light sheet fluorescence microscopy to produce large datasets of the highly resolved cerebrovasculature. Using this data, we trained support vector machines to classify virtual NMR voxels with different dimensions based on the simulated spin dephasing accountable to field inhomogeneities caused by the underlying vasculature. In prediction tests with previously blinded virtual voxels from healthy brain tissue and GL261 tumors, stable classification accuracies above 95% were reached. Our results indicate that high classification accuracies can be stably attained with achievable training set sizes and that larger MRI voxels facilitated increasingly successful classifications, even with small training datasets. We were able to prove that, theoretically, the transverse relaxation process can be harnessed to learn endogenous contrasts for single voxel tissue type classifications on tailored MRI acquisitions. If translatable to experimental MRI, this may augment diagnostic imaging in oncology with automated voxel-by-voxel signal interpretation to detect vascular pathologies.
中文翻译:

基于真实血管诱导自旋相移的虚拟核磁共振体素脑肿瘤分类使用支持向量机
组织微血管系统的重塑通常会促进肿瘤的生长;然而,目前还没有肿瘤学中的成像方式可以无创地量化临床常规中的微血管变化。尽管在典型的磁共振成像 (MRI) 测量中无法解析毛细血管,但它们的几何形状和分布会影响来自每个宏观 MRI 体素的积分核磁共振 (NMR) 信号。我们基于健康和荷瘤小鼠大脑(U87 和 GL261 胶质母细胞瘤)中的真实微血管系统,对不同维度的 NMR 体素中的预期横向弛豫进行了数值模拟。使用光片荧光显微镜获得整个未解剖大脑中的 3D 毛细血管结构,以生成高分辨率脑血管系统的大型数据集。使用这些数据,我们训练了支持向量机,以基于模拟自旋相移对不同维度的虚拟 NMR 体素进行分类,该自旋相移对由底层脉管系统引起的场不均匀性负责。在对来自健康脑组织和 GL261 肿瘤的先前失明的虚拟体素进行的预测测试中,达到了 95% 以上的稳定分类准确度。我们的结果表明,可以通过可实现的训练集大小稳定地获得高分类精度,并且更大的 MRI 体素促进了越来越成功的分类,即使是小的训练数据集也是如此。我们能够证明,理论上,可以利用横向松弛过程来学习定制 MRI 采集中单个体素组织类型分类的内源性对比。如果可以转化为实验性 MRI,
更新日期:2020-04-14
中文翻译:

基于真实血管诱导自旋相移的虚拟核磁共振体素脑肿瘤分类使用支持向量机
组织微血管系统的重塑通常会促进肿瘤的生长;然而,目前还没有肿瘤学中的成像方式可以无创地量化临床常规中的微血管变化。尽管在典型的磁共振成像 (MRI) 测量中无法解析毛细血管,但它们的几何形状和分布会影响来自每个宏观 MRI 体素的积分核磁共振 (NMR) 信号。我们基于健康和荷瘤小鼠大脑(U87 和 GL261 胶质母细胞瘤)中的真实微血管系统,对不同维度的 NMR 体素中的预期横向弛豫进行了数值模拟。使用光片荧光显微镜获得整个未解剖大脑中的 3D 毛细血管结构,以生成高分辨率脑血管系统的大型数据集。使用这些数据,我们训练了支持向量机,以基于模拟自旋相移对不同维度的虚拟 NMR 体素进行分类,该自旋相移对由底层脉管系统引起的场不均匀性负责。在对来自健康脑组织和 GL261 肿瘤的先前失明的虚拟体素进行的预测测试中,达到了 95% 以上的稳定分类准确度。我们的结果表明,可以通过可实现的训练集大小稳定地获得高分类精度,并且更大的 MRI 体素促进了越来越成功的分类,即使是小的训练数据集也是如此。我们能够证明,理论上,可以利用横向松弛过程来学习定制 MRI 采集中单个体素组织类型分类的内源性对比。如果可以转化为实验性 MRI,































 京公网安备 11010802027423号
京公网安备 11010802027423号