当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
NormAE: Deep Adversarial Learning Model to Remove Batch Effects in Liquid Chromatography Mass Spectrometry-Based Metabolomics Data.
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-03-24 , DOI: 10.1021/acs.analchem.9b05460 Zhiwei Rong 1 , Qilong Tan 1 , Lei Cao 1 , Liuchao Zhang 1 , Kui Deng 1 , Yue Huang 1 , Zheng-Jiang Zhu 2 , Zhenzi Li 1 , Kang Li 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-03-24 , DOI: 10.1021/acs.analchem.9b05460 Zhiwei Rong 1 , Qilong Tan 1 , Lei Cao 1 , Liuchao Zhang 1 , Kui Deng 1 , Yue Huang 1 , Zheng-Jiang Zhu 2 , Zhenzi Li 1 , Kang Li 1
Affiliation
|
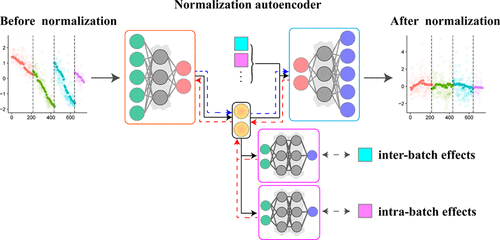
Untargeted metabolomics based on liquid chromatography–mass spectrometry is affected by nonlinear batch effects, which cover up biological effects, result in nonreproducibility, and are difficult to be calibrate. In this study, we propose a novel deep learning model, called Normalization Autoencoder (NormAE), which is based on nonlinear autoencoders (AEs) and adversarial learning. An additional classifier and ranker are trained to provide adversarial regularization during the training of the AE model, latent representations are extracted by the encoder, and then the decoder reconstructs the data without batch effects. The NormAE method was tested on two real metabolomics data sets. After calibration by NormAE, the quality control samples (QCs) for both data sets gathered most closely in a PCA score plot (average distances decreased from 56.550 and 52.476 to 7.383 and 14.075, respectively) and obtained the highest average correlation coefficients (from 0.873 and 0.907 to 0.997 for both). Additionally, NormAE significantly improved biomarker discovery (median number of differential peaks increased from 322 and 466 to 1140 and 1622, respectively). NormAE was compared with four commonly used batch effect removal methods. The results demonstrated that using NormAE produces the best calibration results.
中文翻译:

NormAE:深度对抗学习模型,可消除基于液相色谱质谱的代谢组学数据中的批次效应。
基于液相色谱-质谱法的非靶向代谢组学受非线性批量效应的影响,非线性效应掩盖了生物学效应,导致不可重复性,并且难以校准。在这项研究中,我们提出了一种新型的深度学习模型,称为标准化自动编码器(NormAE),该模型基于非线性自动编码器(AE)和对抗性学习。在AE模型的训练过程中,对其他分类器和等级进行了训练以提供对抗性正则化,编码器提取了潜在表示,然后解码器重建了没有批处理效果的数据。在两个真实的代谢组学数据集上测试了NormAE方法。在通过NormAE进行校准之后,这两个数据集的质量控制样本(QC)在PCA评分图中的收集最为紧密(平均距离从56减少了)。分别为550和52.476到7.383和14.075),并获得了最高的平均相关系数(两者均从0.873和0.907到0.997)。此外,NormAE大大改善了生物标志物的发现(差异峰的中位数分别从322和466增加到1140和1622)。将NormAE与四种常用的批量效应去除方法进行了比较。结果表明,使用NormAE可获得最佳的校准结果。
更新日期:2020-03-24
中文翻译:

NormAE:深度对抗学习模型,可消除基于液相色谱质谱的代谢组学数据中的批次效应。
基于液相色谱-质谱法的非靶向代谢组学受非线性批量效应的影响,非线性效应掩盖了生物学效应,导致不可重复性,并且难以校准。在这项研究中,我们提出了一种新型的深度学习模型,称为标准化自动编码器(NormAE),该模型基于非线性自动编码器(AE)和对抗性学习。在AE模型的训练过程中,对其他分类器和等级进行了训练以提供对抗性正则化,编码器提取了潜在表示,然后解码器重建了没有批处理效果的数据。在两个真实的代谢组学数据集上测试了NormAE方法。在通过NormAE进行校准之后,这两个数据集的质量控制样本(QC)在PCA评分图中的收集最为紧密(平均距离从56减少了)。分别为550和52.476到7.383和14.075),并获得了最高的平均相关系数(两者均从0.873和0.907到0.997)。此外,NormAE大大改善了生物标志物的发现(差异峰的中位数分别从322和466增加到1140和1622)。将NormAE与四种常用的批量效应去除方法进行了比较。结果表明,使用NormAE可获得最佳的校准结果。

































 京公网安备 11010802027423号
京公网安备 11010802027423号