当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In vitro ribosome synthesis and evolution through ribosome display.
Nature Communications ( IF 14.7 ) Pub Date : 2020-02-28 , DOI: 10.1038/s41467-020-14705-2 Michael J Hammerling 1 , Brian R Fritz 1 , Danielle J Yoesep 1 , Do Soon Kim 1 , Erik D Carlson 1, 2 , Michael C Jewett 1, 3, 4, 5, 6
Nature Communications ( IF 14.7 ) Pub Date : 2020-02-28 , DOI: 10.1038/s41467-020-14705-2 Michael J Hammerling 1 , Brian R Fritz 1 , Danielle J Yoesep 1 , Do Soon Kim 1 , Erik D Carlson 1, 2 , Michael C Jewett 1, 3, 4, 5, 6
Affiliation
|
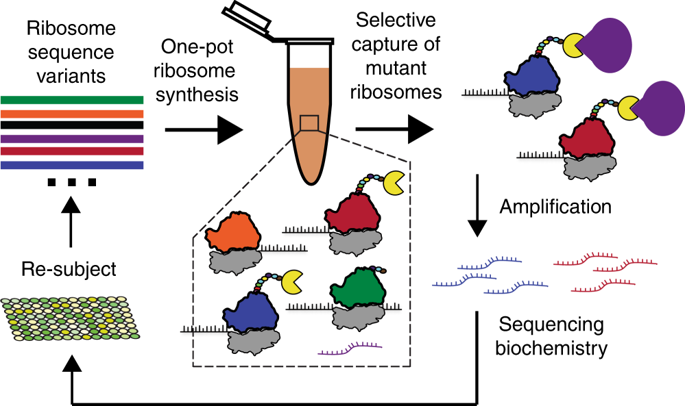
Directed evolution of the ribosome for expanded substrate incorporation and novel functions is challenging because the requirement of cell viability limits the mutations that can be made. Here we address this challenge by combining cell-free synthesis and assembly of translationally competent ribosomes with ribosome display to develop a fully in vitro methodology for ribosome synthesis and evolution (called RISE). We validate the RISE method by selecting active genotypes from a ~1.7 × 107 member library of ribosomal RNA (rRNA) variants, as well as identifying mutant ribosomes resistant to the antibiotic clindamycin from a library of ~4 × 103 rRNA variants. We further demonstrate the prevalence of positive epistasis in resistant genotypes, highlighting the importance of such interactions in selecting for new function. We anticipate that RISE will facilitate understanding of molecular translation and enable selection of ribosomes with altered properties.
中文翻译:

通过核糖体展示的体外核糖体合成和进化。
核糖体定向进化以扩大底物掺入和新功能具有挑战性,因为细胞活力的要求限制了可以进行的突变。在这里,我们通过结合核糖体展示的无细胞合成和翻译能力强大的核糖体组装来解决这一挑战,以开发出一种完整的体外方法进行核糖体合成和进化(称为RISE)。我们通过从〜1.7×107核糖体RNA(rRNA)变体成员库中选择活性基因型,以及从〜4×103 rRNA变体文库中鉴定出对抗生素克林霉素具有抗性的突变核糖体,来验证RISE方法。我们进一步证明了耐药基因型中阳性上位性的普遍性,突出了这种相互作用在选择新功能中的重要性。
更新日期:2020-02-28
中文翻译:

通过核糖体展示的体外核糖体合成和进化。
核糖体定向进化以扩大底物掺入和新功能具有挑战性,因为细胞活力的要求限制了可以进行的突变。在这里,我们通过结合核糖体展示的无细胞合成和翻译能力强大的核糖体组装来解决这一挑战,以开发出一种完整的体外方法进行核糖体合成和进化(称为RISE)。我们通过从〜1.7×107核糖体RNA(rRNA)变体成员库中选择活性基因型,以及从〜4×103 rRNA变体文库中鉴定出对抗生素克林霉素具有抗性的突变核糖体,来验证RISE方法。我们进一步证明了耐药基因型中阳性上位性的普遍性,突出了这种相互作用在选择新功能中的重要性。































 京公网安备 11010802027423号
京公网安备 11010802027423号