当前位置:
X-MOL 学术
›
Angew. Chem. Int. Ed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure of a Protein-RNA Complex by Solid-State NMR Spectroscopy.
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2020-02-28 , DOI: 10.1002/anie.201915465 Mumdooh Ahmed 1 , Alexander Marchanka 1 , Teresa Carlomagno 1, 2
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2020-02-28 , DOI: 10.1002/anie.201915465 Mumdooh Ahmed 1 , Alexander Marchanka 1 , Teresa Carlomagno 1, 2
Affiliation
|
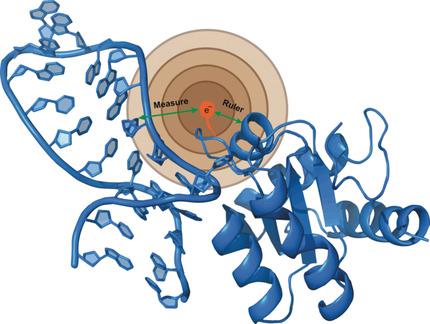
Solid-state NMR (ssNMR) is applicable to high molecular-weight (MW) protein assemblies in a non-amorphous precipitate. The technique yields atomic resolution structural information on both soluble and insoluble particles without limitations of MW or requirement of crystals. Herein, we propose and demonstrate an approach that yields the structure of protein-RNA complexes (RNP) solely from ssNMR data. Instead of using low-sensitivity magnetization transfer steps between heteronuclei of the protein and the RNA, we measure paramagnetic relaxation enhancement effects elicited on the RNA by a paramagnetic tag coupled to the protein. We demonstrate that this data, together with chemical-shift-perturbation data, yields an accurate structure of an RNP complex, starting from the bound structures of its components. The possibility of characterizing protein-RNA interactions by ssNMR may enable applications to large RNP complexes, whose structures are not accessible by other methods.
中文翻译:

通过固态NMR光谱分析蛋白质-RNA复合物的结构。
固态NMR(ssNMR)适用于非非晶形沉淀物中的高分子量(MW)蛋白装配。该技术可在可溶和不可溶颗粒上产生原子分辨率的结构信息,而不受分子量或晶体需求的限制。在本文中,我们提出并证明了仅从ssNMR数据得出蛋白质-RNA复合物(RNP)结构的方法。代替在蛋白质和RNA的杂核之间使用低灵敏度的磁化转移步骤,我们测量与蛋白质偶联的顺磁性标签对RNA引起的顺磁性弛豫增强效果。我们证明,该数据与化学位移扰动数据一起,从其组分的结合结构开始,产生了RNP配合物的准确结构。
更新日期:2020-02-28
中文翻译:

通过固态NMR光谱分析蛋白质-RNA复合物的结构。
固态NMR(ssNMR)适用于非非晶形沉淀物中的高分子量(MW)蛋白装配。该技术可在可溶和不可溶颗粒上产生原子分辨率的结构信息,而不受分子量或晶体需求的限制。在本文中,我们提出并证明了仅从ssNMR数据得出蛋白质-RNA复合物(RNP)结构的方法。代替在蛋白质和RNA的杂核之间使用低灵敏度的磁化转移步骤,我们测量与蛋白质偶联的顺磁性标签对RNA引起的顺磁性弛豫增强效果。我们证明,该数据与化学位移扰动数据一起,从其组分的结合结构开始,产生了RNP配合物的准确结构。































 京公网安备 11010802027423号
京公网安备 11010802027423号