Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Defining the Environmental Adaptations of Genus Devosia: Insights into its Expansive Short Peptide Transport System and Positively Selected Genes.
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41598-020-58163-8 Chandni Talwar 1 , Shekhar Nagar 1 , Roshan Kumar 2 , Joy Scaria 3, 4 , Rup Lal 5 , Ram Krishan Negi 1
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41598-020-58163-8 Chandni Talwar 1 , Shekhar Nagar 1 , Roshan Kumar 2 , Joy Scaria 3, 4 , Rup Lal 5 , Ram Krishan Negi 1
Affiliation
|
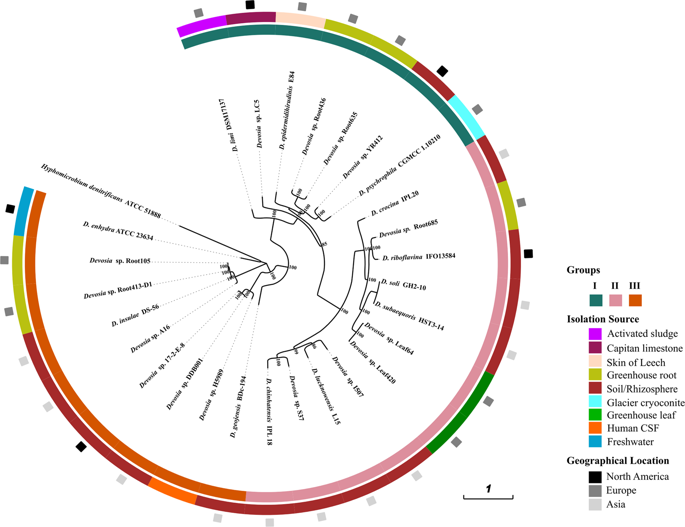
Devosia are well known for their dominance in soil habitats contaminated with various toxins and are best characterized for their bioremediation potential. In this study, we compared the genomes of 27 strains of Devosia with aim to understand their metabolic abilities. The analysis revealed their adaptive gene repertoire which was bared from 52% unique pan-gene content. A striking feature of all genomes was the abundance of oligo- and di-peptide permeases (oppABCDF and dppABCDF) with each genome harboring an average of 60.7 ± 19.1 and 36.5 ± 10.6 operon associated genes respectively. Apart from their primary role in nutrition, these permeases may help Devosia to sense environmental signals and in chemotaxis at stressed habitats. Through sequence similarity network analyses, we identified 29 Opp and 19 Dpp sequences that shared very little homology with any other sequence suggesting an expansive short peptidic transport system within Devosia. The substrate determining components of these permeases viz. OppA and DppA further displayed a large diversity that separated into 12 and 9 homologous clusters respectively in addition to large number of isolated nodes. We also dissected the genome scale positive evolution and found genes associated with growth (exopolyphosphatase, HesB_IscA_SufA family protein), detoxification (moeB, nifU-like domain protein, alpha/beta hydrolase), chemotaxis (cheB, luxR) and stress response (phoQ, uspA, luxR, sufE) were positively selected. The study highlights the genomic plasticity of the Devosia spp. for conferring adaptation, bioremediation and the potential to utilize a wide range of substrates. The widespread toxin-antitoxin loci and 'open' state of the pangenome provided evidence of plastic genomes and a much larger genetic repertoire of the genus which is yet uncovered.
中文翻译:

定义灵芝属植物的环境适应性:深入了解其广泛的短肽转运系统和积极选择的基因。
德沃西亚以其在被各种毒素污染的土壤生境中的优势而著称,并以其生物修复潜力为特征。在这项研究中,我们比较了27种Devosia菌株的基因组,以了解其代谢能力。分析显示他们的适应性基因库被限制在52%的独特泛基因含量之内。所有基因组的一个显着特征是寡肽和二肽通透酶(oppABCDF和dppABCDF)的丰度,每个基因组分别带有平均60.7±19.1和36.5±10.6操纵子相关基因。除了它们在营养中的主要作用外,这些通透酶还可以帮助Devosia感知环境信号以及在紧张的栖息地进行趋化性。通过序列相似性网络分析,我们确定了29个Opp和19个Dpp序列,这些序列与任何其他序列的同源性极低,这表明Devosia中存在着广泛的短肽转运系统。这些渗透酶的底物确定成分即。OppA和DppA进一步显示出很大的多样性,除了有大量的孤立节点,它们还分别分为12和9个同源簇。我们还解剖了基因组规模的正向进化过程,并发现了与生长相关的基因(胞外磷酸酶,HesB_IscA_SufA家族蛋白),排毒(moeB,nifU样结构域蛋白,α/β水解酶),趋化性(cheB,luxR)和应激反应(phoQ,肯定选择了uspA,luxR,sufE)。该研究突出了Devosia spp的基因组可塑性。为了赋予适应性,生物修复以及利用多种底物的潜力。广泛的毒素-抗毒素基因座和泛基因组的“开放”状态提供了可塑性基因组的证据,以及该基因尚未发现的更大的遗传库。
更新日期:2020-01-24
中文翻译:

定义灵芝属植物的环境适应性:深入了解其广泛的短肽转运系统和积极选择的基因。
德沃西亚以其在被各种毒素污染的土壤生境中的优势而著称,并以其生物修复潜力为特征。在这项研究中,我们比较了27种Devosia菌株的基因组,以了解其代谢能力。分析显示他们的适应性基因库被限制在52%的独特泛基因含量之内。所有基因组的一个显着特征是寡肽和二肽通透酶(oppABCDF和dppABCDF)的丰度,每个基因组分别带有平均60.7±19.1和36.5±10.6操纵子相关基因。除了它们在营养中的主要作用外,这些通透酶还可以帮助Devosia感知环境信号以及在紧张的栖息地进行趋化性。通过序列相似性网络分析,我们确定了29个Opp和19个Dpp序列,这些序列与任何其他序列的同源性极低,这表明Devosia中存在着广泛的短肽转运系统。这些渗透酶的底物确定成分即。OppA和DppA进一步显示出很大的多样性,除了有大量的孤立节点,它们还分别分为12和9个同源簇。我们还解剖了基因组规模的正向进化过程,并发现了与生长相关的基因(胞外磷酸酶,HesB_IscA_SufA家族蛋白),排毒(moeB,nifU样结构域蛋白,α/β水解酶),趋化性(cheB,luxR)和应激反应(phoQ,肯定选择了uspA,luxR,sufE)。该研究突出了Devosia spp的基因组可塑性。为了赋予适应性,生物修复以及利用多种底物的潜力。广泛的毒素-抗毒素基因座和泛基因组的“开放”状态提供了可塑性基因组的证据,以及该基因尚未发现的更大的遗传库。






























 京公网安备 11010802027423号
京公网安备 11010802027423号