Journal of Proteomics ( IF 2.8 ) Pub Date : 2020-01-24 , DOI: 10.1016/j.jprot.2020.103669 Lei Zhao 1 , Xiaoji Cong 2 , Linhui Zhai 3 , Hao Hu 3 , Jun-Yu Xu 3 , Wensi Zhao 2 , Mengdi Zhu 2 , Minjia Tan 2 , Bang-Ce Ye 1
|
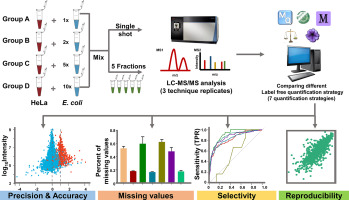
The selection of a data processing method for use in mass spectrometry-based label-free proteome quantification contributes significantly to its accuracy and precision. In this study, we comprehensively evaluated 7 commonly-used label-free quantification methods (MaxQuant-Spectrum count, MaxQuant-iBAQ, MaxQuant-LFQ, MaxQuant-LFAQ, Proteome Discoverer, MetaMorpheus, TPP-StPeter) with a focus on missing values, precision, accuracy, selectivity, and reproducibility of low abundance protein quantification in both single shot and fractionation. Our results showed that among the tested strategies, MaxQuant in MaxLFQ mode outperformed other strategies in terms of accuracy and precision in both whole proteome and low abundance proteome quantification, whereas the Proteome Discoverer (PD) strategy using SEQUEST as a search engine performed better in terms of quantifiable low abundance proteome coverage. We subsequently applied the PD and MaxLFQ strategies in a blood proteomic dataset and found that many FDA-approved tumor prognostic biomarkers could be identified as well as quantified using the PD strategy, indicating the potential advantage of PD in label-free quantification studies. These results provide a reference for method choice in label-free quantification data analysis.
Significance
Mass spectrometry-based label-free quantification methods play an important role in label-free proteome data analysis. In this study, we evaluated 7 commonly-used label-free quantification methods with respect to the following aspects: missing values, precision, accuracy, selectivity, and reproducibility for low abundance protein quantification. The results showed that, among the strategies evaluated, the PD strategy with SEQUEST as a search engine performed better in terms of low abundance protein coverage. This study provides a reference for method choice in label-free quantification data analysis.
中文翻译:

无标签量化策略的比较评估。
选择用于基于质谱的无标记蛋白质组定量的数据处理方法将极大地提高其准确性和准确性。在这项研究中,我们全面评估了7种常用的无标记定量方法(MaxQuant-Spectrum计数,MaxQuant-iBAQ,MaxQuant-LFQ,MaxQuant-LFAQ,Proteome Discoverer,MetaMorpheus,TPP-StPeter),重点是缺失值,单次注射和分馏中低丰度蛋白质定量的精密度,准确性,选择性和重现性。我们的结果表明,在测试的策略中,MaxLFQ模式下的MaxQuant在整个蛋白质组和低丰度蛋白质组定量方面的准确性和精密度均优于其他策略,而使用SEQUEST作为搜索引擎的蛋白质组发现器(PD)策略在可量化的低丰度蛋白质组覆盖方面表现更好。我们随后在血液蛋白质组学数据集中应用了PD和MaxLFQ策略,发现可以通过PD策略识别和定量许多FDA批准的肿瘤预后生物标志物,这表明PD在无标记定量研究中的潜在优势。这些结果为无标记定量数据分析中方法的选择提供了参考。表明PD在无标记定量研究中的潜在优势。这些结果为无标记定量数据分析中方法的选择提供了参考。表明PD在无标记定量研究中的潜在优势。这些结果为无标记定量数据分析中方法的选择提供了参考。
意义
基于质谱的无标记定量方法在无标记蛋白质组数据分析中起着重要作用。在这项研究中,我们针对以下方面评估了7种常用的无标记定量方法:缺失值,准确性,准确性,选择性和低丰度蛋白质定量的可再现性。结果表明,在评估的策略中,以SEQUEST作为搜索引擎的PD策略在低丰度蛋白质覆盖率方面表现更好。本研究为无标签定量数据分析中方法的选择提供了参考。

































 京公网安备 11010802027423号
京公网安备 11010802027423号