当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize.
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-07 , DOI: 10.1038/s41467-019-14023-2 Changsheng Li 1, 2 , Xiaoli Xiang 2, 3 , Yongcai Huang 2, 4 , Yong Zhou 2 , Dong An 1 , Jiaqiang Dong 5 , Chenxi Zhao 6 , Hongjun Liu 7 , Yubin Li 8 , Qiong Wang 2 , Chunguang Du 9 , Joachim Messing 5 , Brian A Larkins 10 , Yongrui Wu 2 , Wenqin Wang 1
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-07 , DOI: 10.1038/s41467-019-14023-2 Changsheng Li 1, 2 , Xiaoli Xiang 2, 3 , Yongcai Huang 2, 4 , Yong Zhou 2 , Dong An 1 , Jiaqiang Dong 5 , Chenxi Zhao 6 , Hongjun Liu 7 , Yubin Li 8 , Qiong Wang 2 , Chunguang Du 9 , Joachim Messing 5 , Brian A Larkins 10 , Yongrui Wu 2 , Wenqin Wang 1
Affiliation
|
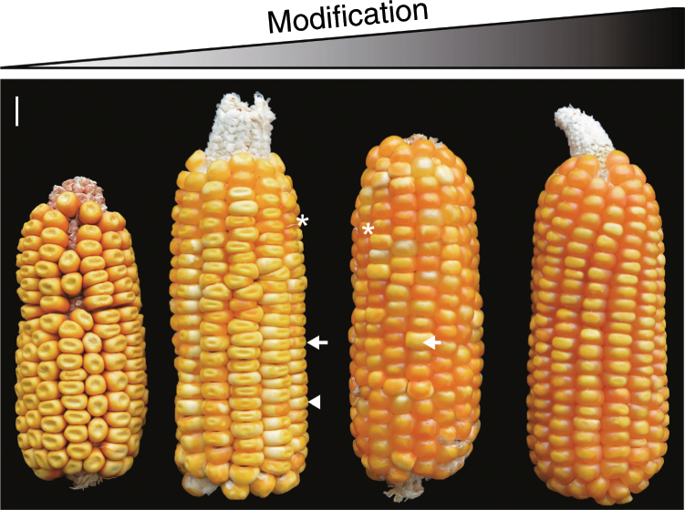
Mutation of o2 doubles maize endosperm lysine content, but it causes an inferior kernel phenotype. Developing quality protein maize (QPM) by introgressing o2 modifiers (Mo2s) into the o2 mutant benefits millions of people in developing countries where maize is a primary protein source. Here, we report genome sequence and annotation of a South African QPM line K0326Y, which is assembled from single-molecule, real-time shotgun sequencing reads collinear with an optical map. We achieve a N50 contig length of 7.7 million bases (Mb) directly from long-read assembly, compared to those of 1.04 Mb for B73 and 1.48 Mb for Mo17. To characterize Mo2s, we map QTLs to chromosomes 1, 6, 7, and 9 using an F2 population derived from crossing K0326Y and W64Ao2. RNA-seq analysis of QPM and o2 endosperms reveals a group of differentially expressed genes that coincide with Mo2 QTLs, suggesting a potential role in vitreous endosperm formation.
更新日期:2020-01-07































 京公网安备 11010802027423号
京公网安备 11010802027423号