Mini-Reviews in Medicinal Chemistry ( IF 3.3 ) Pub Date : 2020-06-30 , DOI: 10.2174/1389557519666190904151227 Deepika Purohit 1 , Vandana Saini 2 , Sanjiv Kumar 1 , Ajit Kumar 2 , Balasubramanian Narasimhan 1
|
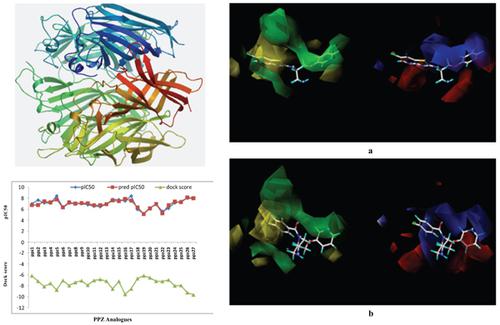
Background & Objective: Comparative molecular field analysis (CoMFA) of 27 analogues of 2-((pyridin-3-yloxy)methyl)piperazine derivatives was carried out using software Tripos SYBYL X. Optimal r2 (0.854) and q2 (0.541) values were obtained for the developed 3D-QSAR model. The contour plots obtained from CoMFA analysis have shown 13.84% steric contribution and 66.14% electrostatic contribution towards an anti-inflammatory activity.
Methods: The homology model of the receptor protein, α7 nicotinic acetylcholine, was generated in SWISS MODELLER using auto template mode and was analysed for the quality using Procheck, QMEAN Z-score, Anolea and GROMOS plots. The QMEAN score for the model was observed to be - 3.862. The generated model of alpha 7 nicotinic acetylcholine receptor was used for docking study of 27 piperazine analogues using Auto-Dock 4.2.5.1.
Results: The dock score obtained from docking analysis was then correlated with experimental pIC50 values for in-silico validation of the developed CoMFA model and a good correlation was obtained with correlation coefficient (r2) value of -0.7378.
Conclusion: The present investigation suggests an optimal 3D-QSAR with CoMFA model for further evaluating new chemical entities based on piperazine skeleton.
中文翻译:

2-((吡啶-3-基氧基)甲基)哌嗪作为α7烟碱乙酰胆碱受体调节剂的三维定量构效关系(3DQSAR)和分子对接研究,用于治疗炎症性疾病。
背景与目的:使用Tripos SYBYL X软件对27种2-((吡啶-3-基氧基)甲基)哌嗪衍生物的类似物进行比较分子场分析(CoMFA)。最佳r2(0.854)和q2(0.541)值为为开发的3D-QSAR模型获得。从CoMFA分析获得的轮廓图显示,抗炎活性对空间的贡献为13.84%,对静电的贡献为66.14%。
方法:使用自动模板模式在SWISS MODELLER中生成受体蛋白α7烟碱乙酰胆碱的同源性模型,并使用Procheck,QMEAN Z评分,Anolea和GROMOS图对质量进行分析。该模型的QMEAN得分为-3.862。使用Auto-Dock 4.2.5.1,将生成的α7烟碱乙酰胆碱受体模型用于27个哌嗪类似物的对接研究。
结果:通过对接分析获得的对接得分随后与实验pIC50值相关联,以对开发的CoMFA模型进行计算机内验证,并获得了很好的相关性,相关系数(r2)值为-0.7378。
结论:目前的研究表明,使用CoMFA模型的最佳3D-QSAR可进一步评估基于哌嗪骨架的新化学实体。











































 京公网安备 11010802027423号
京公网安备 11010802027423号