当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Highly Selective and Sensitive Electrochemiluminescence Biosensor for p53 DNA Sequence Based on Nicking Endonuclease Assisted Target Recycling and Hyperbranched Rolling Circle Amplification
Analytical Chemistry ( IF 6.7 ) Pub Date : 2016-04-27 00:00:00 , DOI: 10.1021/acs.analchem.5b04521 Linlin Yang 1 , Yingzhou Tao 1 , Guiyin Yue 1 , Ruibao Li 1 , Bin Qiu 1 , Longhua Guo 1 , Zhenyu Lin 1 , Huang-Hao Yang 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2016-04-27 00:00:00 , DOI: 10.1021/acs.analchem.5b04521 Linlin Yang 1 , Yingzhou Tao 1 , Guiyin Yue 1 , Ruibao Li 1 , Bin Qiu 1 , Longhua Guo 1 , Zhenyu Lin 1 , Huang-Hao Yang 1
Affiliation
|
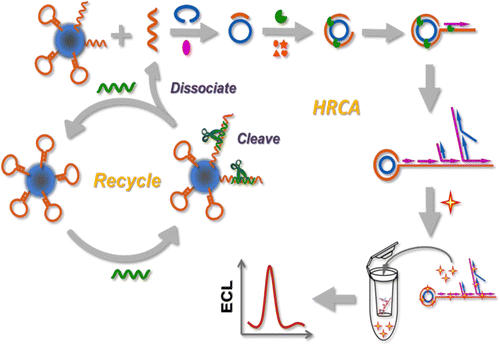
An ultrasensitive and specific electrochemiluminescence (ECL) biosensor has been designed for the p53 DNA sequence, which is based on cascade signal amplification of nicking endonuclease assisted target recycling and hyperbranched rolling circle amplification (HRCA). First of all, biotin modified hairpin capture DNA (HP) probe was immobilized on the surface of streptavidin magnespheres paramagnetic particles (PMPs). Target DNA hybridized with the loop portion of the HP probe, therefore unfolding HP to form a double-stranded DNA (dsDNA) containing the specific nicking site of the nicking endonuclease. Then, the nicking endonuclease recognized the specific nicking site and cleaved the HP into two pieces, liberating target DNA and the complementary sequence piece for the padlock probe. The intact target DNA would initiate the next cycle of hybridization and cleavage, thereby releasing multiple complementary sequences for the padlock probes. The liberated complementary sequences hybridized with the padlock probes, subsequently inducing the HRCA reaction and generating numerous dsDNA segments. Herein, Ru(phen)32+ was embedded into dsDNA and worked as ECL signal reporter. The reaction products were eventually pretreated by dialysis tube with the cutoff membrane to remove the residual Ru(phen)32+ in the solution for the following ECL measurements. Using this cascade amplification strategy, an ultrasensitive p53 DNA sequence detection method was developed with a wide linear range from 0.05 to 100 fM and a low detection limit of 0.02 fM. Moreover, this cascade amplified ECL biosensor had specific recognition capacity for noncomplementary and single- and double-base mismatched DNA. The proposed ECL biosensor might have a great potential in biomedical research and clinic analysis.
中文翻译:

基于切口核酸内切酶辅助靶点回收和超支化滚环扩增的p53 DNA序列的高选择性和灵敏的电化学发光生物传感器
针对p53 DNA序列设计了一种超灵敏的特异性电致发光(ECL)生物传感器,该传感器基于切口内切核酸酶辅助靶标回收的级联信号扩增和超支化滚环扩增(HRCA)。首先,将生物素修饰的发夹捕获DNA(HP)探针固定在链霉亲和素磁珠顺磁性颗粒(PMP)的表面上。目标DNA与HP探针的环部分杂交,因此展开HP,形成包含切口核酸内切酶特定切口位点的双链DNA(dsDNA)。然后,切口内切核酸酶识别了特定的切口位点,并将HP切割成两段,从而释放了目标DNA和用于挂锁探针的互补序列。完整的靶DNA将启动杂交和切割的下一个周期,从而释放出多个用于挂锁探针的互补序列。释放的互补序列与挂锁探针杂交,随后诱导HRCA反应并生成许多dsDNA片段。在此,Ru(phen)3 2+被嵌入dsDNA,并作为ECL信号报告基因。最终,用截留膜通过透析管对反应产物进行预处理,以除去溶液中残留的Ru(phen)3 2+,以进行以下ECL测量。使用这种级联扩增策略,开发了一种超灵敏的p53 DNA序列检测方法,其线性范围从0.05到100 fM,检测下限为0.02 fM。此外,这种级联扩增的ECL生物传感器对非互补和单碱基和双碱基错配的DNA具有特定的识别能力。提出的ECL生物传感器在生物医学研究和临床分析中可能具有巨大的潜力。
更新日期:2016-04-27
中文翻译:

基于切口核酸内切酶辅助靶点回收和超支化滚环扩增的p53 DNA序列的高选择性和灵敏的电化学发光生物传感器
针对p53 DNA序列设计了一种超灵敏的特异性电致发光(ECL)生物传感器,该传感器基于切口内切核酸酶辅助靶标回收的级联信号扩增和超支化滚环扩增(HRCA)。首先,将生物素修饰的发夹捕获DNA(HP)探针固定在链霉亲和素磁珠顺磁性颗粒(PMP)的表面上。目标DNA与HP探针的环部分杂交,因此展开HP,形成包含切口核酸内切酶特定切口位点的双链DNA(dsDNA)。然后,切口内切核酸酶识别了特定的切口位点,并将HP切割成两段,从而释放了目标DNA和用于挂锁探针的互补序列。完整的靶DNA将启动杂交和切割的下一个周期,从而释放出多个用于挂锁探针的互补序列。释放的互补序列与挂锁探针杂交,随后诱导HRCA反应并生成许多dsDNA片段。在此,Ru(phen)3 2+被嵌入dsDNA,并作为ECL信号报告基因。最终,用截留膜通过透析管对反应产物进行预处理,以除去溶液中残留的Ru(phen)3 2+,以进行以下ECL测量。使用这种级联扩增策略,开发了一种超灵敏的p53 DNA序列检测方法,其线性范围从0.05到100 fM,检测下限为0.02 fM。此外,这种级联扩增的ECL生物传感器对非互补和单碱基和双碱基错配的DNA具有特定的识别能力。提出的ECL生物传感器在生物医学研究和临床分析中可能具有巨大的潜力。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号