Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transcriptome analysis of Aeromonas hydrophila infected hybrid sturgeon (Huso dauricus×Acipenser schrenckii).
Scientific Reports ( IF 3.8 ) Pub Date : 2018-Dec-18 , DOI: 10.1038/s41598-018-36376-2 Nan Jiang , Yuding Fan , Yong Zhou , Weiling Wang , Jie Ma , Lingbing Zeng
Scientific Reports ( IF 3.8 ) Pub Date : 2018-Dec-18 , DOI: 10.1038/s41598-018-36376-2 Nan Jiang , Yuding Fan , Yong Zhou , Weiling Wang , Jie Ma , Lingbing Zeng
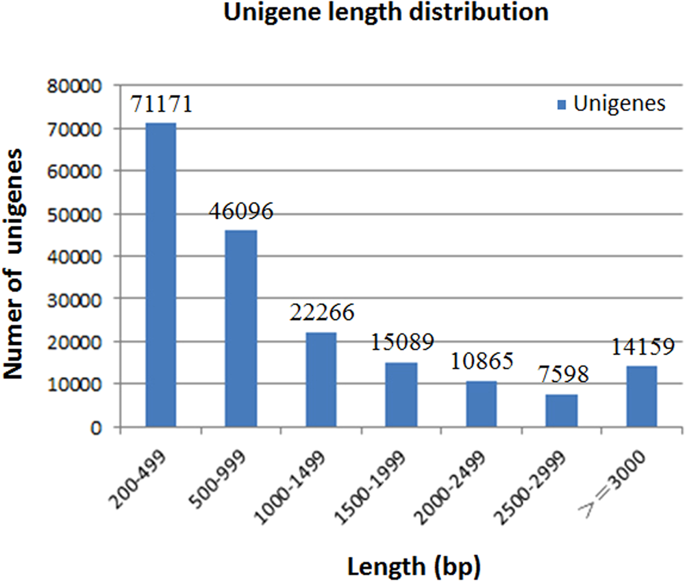
|
The hybrid sturgeon (Huso dauricus × Acipenser schrenckii) is an economically important species in China. With the increasing aquaculture of hybrid sturgeon, the bacterial diseases are a great concern of the industry. In this study, de novo sequencing was used to compare the difference in transcriptome in spleen of the infected and mock infected sturgeon with Aeromonas hydrophila. Among 187,244 unigenes obtained, 87,887 unigenes were annotated and 1,147 unigenes were associated with immune responses genes. Comparative expression analysis indicated that 2,723 differently expressed genes between the infected and mock-infected group were identified, including 1,420 up-regulated and 1,303 down-regulated genes. 283 differently expressed anti-bacterial immune related genes were scrutinized, including 168 up-regulated and 115 down-regulated genes. Ten of the differently expressed genes were further validated by qRT-PCR. In this study, toll like receptors (TLRs) pathway, NF-kappa B pathway, class A scavenger receptor pathway, phagocytosis pathway, mannose receptor pathway and complement pathway were shown to be up-regulated in Aeromonas hydrophila infected hybrid sturgeon. Additionally, 65,040 potential SSRs and 2,133,505 candidate SNPs were identified from the hybrid sturgeon spleen transcriptome. This study could provide an insight of host immune genes associated with bacterial infection in hybrid sturgeon.
更新日期:2018-12-18





















































 京公网安备 11010802027423号
京公网安备 11010802027423号