Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Bioorthogonal protein-DNA conjugation methods for force spectroscopy.
Scientific Reports ( IF 3.8 ) Pub Date : 2019-09-25 , DOI: 10.1038/s41598-019-49843-1 Marie Synakewicz 1 , Daniela Bauer 2 , Matthias Rief 2 , Laura S Itzhaki 1
Scientific Reports ( IF 3.8 ) Pub Date : 2019-09-25 , DOI: 10.1038/s41598-019-49843-1 Marie Synakewicz 1 , Daniela Bauer 2 , Matthias Rief 2 , Laura S Itzhaki 1
Affiliation
|
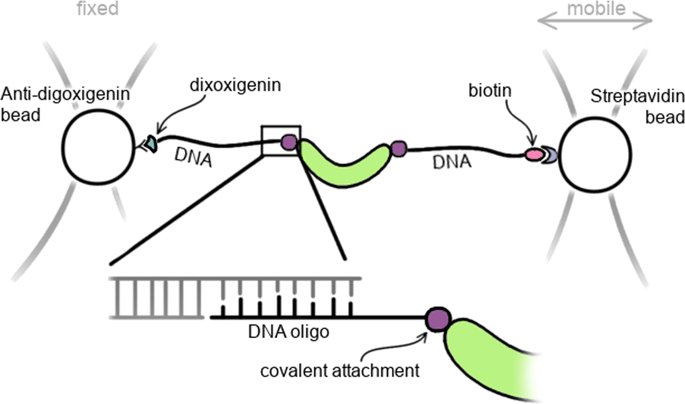
Accurate and stable site-specific attachment of DNA molecules to proteins is a requirement for many single-molecule force spectroscopy techniques. The most commonly used method still relies on maleimide chemistry involving cysteine residues in the protein of interest. Studies have consequently often focused on model proteins that either have no cysteines or with a small number of cysteines that can be deleted so that cysteines can then be introduced at specific sites. However, many proteins, especially in eukaryotes, contain too many cysteine residues to be amenable to this strategy, and therefore there is tremendous need for new and broadly applicable approaches to site-specific conjugation. Here we present bioorthogonal approaches for making DNA-protein conjugates required in force spectroscopy experiments. Unnatural amino acids are introduced site-specifically and conjugated to DNA oligos bearing the respective modifications to undergo either strain-promoted azidealkyne cycloaddition (SPAAC) or inverse-electron-demand Diels-Alder (IE-DA) reactions. We furthermore show that SPAAC is compatible with a previously published peptide-based attachment approach. By expanding the available toolkit to tag-free methods based on bioorthogonal reactions, we hope to enable researchers to interrogate the mechanics of a much broader range of proteins than is currently possible.
中文翻译:

力光谱的生物正交蛋白质-DNA偶联方法。
DNA分子与蛋白质的精确且稳定的位点特异性连接是许多单分子力光谱技术的要求。最常用的方法仍依赖于涉及目标蛋白质中半胱氨酸残基的马来酰亚胺化学。因此,研究通常集中在没有半胱氨酸或具有少量可被删除的半胱氨酸的模型蛋白上,从而可将半胱氨酸引入特定位点。然而,许多蛋白质,特别是在真核生物中,含有太多的半胱氨酸残基,无法适应该策略,因此,迫切需要针对位点特异性缀合的新方法和广泛适用的方法。在这里,我们介绍了用于制备力谱实验所需的DNA-蛋白质共轭物的生物正交方法。非天然氨基酸被定点引入,并与带有相应修饰的DNA寡核苷酸缀合,以进行应变促进的叠氮炔环加成反应(SPAAC)或电子反需求Diels-Alder反应(IE-DA)。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够研究比目前可能的蛋白质范围更广的力学。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够对比目前可能的蛋白质范围更广泛的蛋白质进行研究。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够研究比目前可能的蛋白质范围更广的力学。
更新日期:2019-09-26
中文翻译:

力光谱的生物正交蛋白质-DNA偶联方法。
DNA分子与蛋白质的精确且稳定的位点特异性连接是许多单分子力光谱技术的要求。最常用的方法仍依赖于涉及目标蛋白质中半胱氨酸残基的马来酰亚胺化学。因此,研究通常集中在没有半胱氨酸或具有少量可被删除的半胱氨酸的模型蛋白上,从而可将半胱氨酸引入特定位点。然而,许多蛋白质,特别是在真核生物中,含有太多的半胱氨酸残基,无法适应该策略,因此,迫切需要针对位点特异性缀合的新方法和广泛适用的方法。在这里,我们介绍了用于制备力谱实验所需的DNA-蛋白质共轭物的生物正交方法。非天然氨基酸被定点引入,并与带有相应修饰的DNA寡核苷酸缀合,以进行应变促进的叠氮炔环加成反应(SPAAC)或电子反需求Diels-Alder反应(IE-DA)。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够研究比目前可能的蛋白质范围更广的力学。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够对比目前可能的蛋白质范围更广泛的蛋白质进行研究。我们进一步表明,SPAAC与以前发布的基于肽的附着方法兼容。通过将可用的工具包扩展到基于生物正交反应的无标签方法,我们希望使研究人员能够研究比目前可能的蛋白质范围更广的力学。





















































 京公网安备 11010802027423号
京公网安备 11010802027423号