Nature Protocols ( IF 13.1 ) Pub Date : 2019-03-18 , DOI: 10.1038/s41596-018-0119-1 André Brodkorb , Lotti Egger , Marie Alminger , Paula Alvito , Ricardo Assunção , Simon Ballance , Torsten Bohn , Claire Bourlieu-Lacanal , Rachel Boutrou , Frédéric Carrière , Alfonso Clemente , Milena Corredig , Didier Dupont , Claire Dufour , Cathrina Edwards , Matt Golding , Sibel Karakaya , Bente Kirkhus , Steven Le Feunteun , Uri Lesmes , Adam Macierzanka , Alan R. Mackie , Carla Martins , Sébastien Marze , David Julian McClements , Olivia Ménard , Mans Minekus , Reto Portmann , Cláudia N. Santos , Isabelle Souchon , R. Paul Singh , Gerd E. Vegarud , Martin S. J. Wickham , Werner Weitschies , Isidra Recio
|
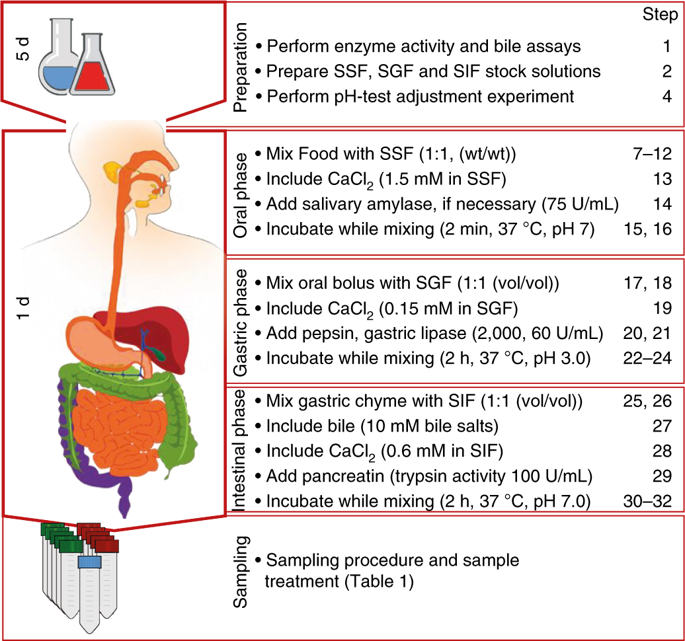
Developing a mechanistic understanding of the impact of food structure and composition on human health has increasingly involved simulating digestion in the upper gastrointestinal tract. These simulations have used a wide range of different conditions that often have very little physiological relevance, and this impedes the meaningful comparison of results. The standardized protocol presented here is based on an international consensus developed by the COST INFOGEST network. The method is designed to be used with standard laboratory equipment and requires limited experience to encourage a wide range of researchers to adopt it. It is a static digestion method that uses constant ratios of meal to digestive fluids and a constant pH for each step of digestion. This makes the method simple to use but not suitable for simulating digestion kinetics. Using this method, food samples are subjected to sequential oral, gastric and intestinal digestion while parameters such as electrolytes, enzymes, bile, dilution, pH and time of digestion are based on available physiological data. This amended and improved digestion method (INFOGEST 2.0) avoids challenges associated with the original method, such as the inclusion of the oral phase and the use of gastric lipase. The method can be used to assess the endpoints resulting from digestion of foods by analyzing the digestion products (e.g., peptides/amino acids, fatty acids, simple sugars) and evaluating the release of micronutrients from the food matrix. The whole protocol can be completed in ~7 d, including ~5 d required for the determination of enzyme activities.
中文翻译:

INFOGEST胃肠道食物消化的静态体外模拟
对食物结构和成分对人类健康的影响的机械理解,越来越多地涉及模拟上消化道的消化。这些模拟使用了很多不同的条件,这些条件通常没有什么生理相关性,这阻碍了结果的有意义的比较。这里介绍的标准化协议基于COST INFOGEST网络开发的国际共识。该方法旨在与标准实验室设备一起使用,并且经验有限,以鼓励广泛的研究人员采用该方法。这是一种静态消化方法,在消化的每个步骤中都使用恒定的粗粉与消化液的比例和恒定的pH值。这使该方法易于使用,但不适用于模拟消化动力学。使用这种方法,可以对食物样品进行连续的口腔,胃和肠消化,而诸如电解质,酶,胆汁,稀释度,pH和消化时间等参数是基于可用的生理数据。这种经过改进的改进的消化方法(INFOGEST 2.0)避免了与原始方法相关的挑战,例如包括口服相和胃脂肪酶的使用。该方法可用于通过分析消化产物(例如肽/氨基酸,脂肪酸,单糖)并评估微量营养素从食物基质中的释放来评估由食物消化产生的终点。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。胃和肠消化,而诸如电解质,酶,胆汁,稀释度,pH和消化时间等参数则基于可用的生理数据。这种经过改进的改进的消化方法(INFOGEST 2.0)避免了与原始方法相关的挑战,例如包括口服相和胃脂肪酶的使用。该方法可用于通过分析消化产物(例如肽/氨基酸,脂肪酸,单糖)并评估微量营养素从食物基质中的释放来评估由食物消化产生的终点。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。胃和肠消化,而诸如电解质,酶,胆汁,稀释度,pH和消化时间等参数则基于可用的生理数据。这种经过改进的改进的消化方法(INFOGEST 2.0)避免了与原始方法相关的挑战,例如包含口服相和使用胃脂肪酶。该方法可用于通过分析消化产物(例如肽/氨基酸,脂肪酸,单糖)并评估微量营养素从食物基质中的释放来评估由食物消化产生的终点。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。这种经过改进的改进的消化方法(INFOGEST 2.0)避免了与原始方法相关的挑战,例如包含口服相和使用胃脂肪酶。该方法可用于通过分析消化产物(例如肽/氨基酸,脂肪酸,单糖)并评估微量营养素从食物基质中的释放来评估由食物消化产生的终点。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。这种经过改进的改进的消化方法(INFOGEST 2.0)避免了与原始方法相关的挑战,例如包含口服相和使用胃脂肪酶。该方法可用于通过分析消化产物(例如肽/氨基酸,脂肪酸,单糖)并评估微量营养素从食物基质中的释放来评估由食物消化产生的终点。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。单糖),并评估食物基质中微量营养素的释放。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。单糖),并评估食物基质中微量营养素的释放。整个方案可在约7天内完成,包括测定酶活性所需的约5天内。











































 京公网安备 11010802027423号
京公网安备 11010802027423号