当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
AlzyFinder: A Machine-Learning-Driven Platform for Ligand-Based Virtual Screening and Network Pharmacology
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-31 , DOI: 10.1021/acs.jcim.4c01481 Jessica Valero-Rojas, Camilo Ramírez-Sánchez, Laura Pacheco-Paternina, Paulina Valenzuela-Hormazabal, Fernanda I. Saldivar-González, Paula Santana, Janneth González, Tatiana Gutiérrez-Bunster, Alejandro Valdés-Jiménez, David Ramírez
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-31 , DOI: 10.1021/acs.jcim.4c01481 Jessica Valero-Rojas, Camilo Ramírez-Sánchez, Laura Pacheco-Paternina, Paulina Valenzuela-Hormazabal, Fernanda I. Saldivar-González, Paula Santana, Janneth González, Tatiana Gutiérrez-Bunster, Alejandro Valdés-Jiménez, David Ramírez
|
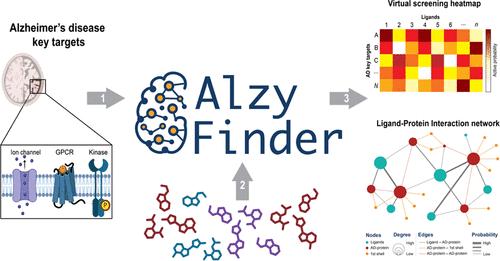
Alzheimer’s disease (AD), a prevalent neurodegenerative disorder, presents significant challenges in drug development due to its multifactorial nature and complex pathophysiology. The AlzyFinder Platform, introduced in this study, addresses these challenges by providing a comprehensive, free web-based tool for parallel ligand-based virtual screening and network pharmacology, specifically targeting over 85 key proteins implicated in AD. This innovative approach is designed to enhance the identification and analysis of potential multitarget ligands, thereby accelerating the development of effective therapeutic strategies against AD. AlzyFinder Platform incorporates machine learning models to facilitate the ligand-based virtual screening process. These models, built with the XGBoost algorithm and optimized through Optuna, were meticulously trained and validated using robust methodologies to ensure high predictive accuracy. Validation included extensive testing with active, inactive, and decoy molecules, demonstrating the platform’s efficacy in distinguishing active compounds. The models are evaluated based on balanced accuracy, precision, and F1 score metrics. A unique soft-voting ensemble approach is utilized to refine the classification process, integrating the strengths of individual models. This methodological framework enables a comprehensive analysis of interaction data, which is presented in multiple formats such as tables, heat maps, and interactive Ligand–Protein Interaction networks, thus enhancing the visualization and analysis of drug–protein interactions. AlzyFinder was applied to screen five molecules recently reported (and not used to train or validate the ML models) as active compounds against five key AD targets. The platform demonstrated its efficacy by accurately predicting all five molecules as true positives with a probability greater than 0.70. This result underscores the platform’s capability in identifying potential therapeutic compounds with high precision. In conclusion, AlzyFinder’s innovative approach extends beyond traditional virtual screening by incorporating network pharmacology analysis, thus providing insights into the systemic actions of drug candidates. This feature allows for the exploration of ligand–protein and protein–protein interactions and their extensions, offering a comprehensive view of potential therapeutic impacts. As the first open-access platform of its kind, AlzyFinder stands as a valuable resource for the AD research community, available at http://www.alzyfinder-platform.udec.cl with supporting data and scripts accessible via GitHub https://github.com/ramirezlab/AlzyFinder.
中文翻译:

AlzyFinder:用于基于配体的虚拟筛选和网络药理学的机器学习驱动平台
阿尔茨海默病 (AD) 是一种普遍存在的神经退行性疾病,由于其多因素性质和复杂的病理生理学,给药物开发带来了重大挑战。本研究中引入的 AlzyFinder 平台通过为基于平行配体的虚拟筛选和网络药理学提供一个全面的、免费的基于 Web 的工具来应对这些挑战,专门针对超过 85 种与 AD 相关的关键蛋白。这种创新方法旨在加强对潜在多靶标配体的识别和分析,从而加速开发针对 AD 的有效治疗策略。AlzyFinder 平台整合了机器学习模型,以促进基于配体的虚拟筛选过程。这些模型使用 XGBoost 算法构建并通过 Optuna 进行优化,使用强大的方法进行精心训练和验证,以确保高预测准确性。验证包括对活性、非活性和诱饵分子进行的广泛测试,证明了该平台在区分活性化合物方面的功效。这些模型根据平衡的准确度、精度和 F1 分数指标进行评估。采用独特的软投票集成方法来优化分类过程,整合各个模型的优势。该方法框架能够对相互作用数据进行全面分析,这些数据以多种格式呈现,例如表格、热图和交互式配体-蛋白质相互作用网络,从而增强药物-蛋白质相互作用的可视化和分析。AlzyFinder 用于筛选最近报道的 5 种分子(未用于训练或验证 ML 模型)作为针对 5 个关键 AD 靶标的活性化合物。 该平台通过准确预测所有五个分子为真阳性(概率大于 0.70)来证明其疗效。这一结果强调了该平台高精度识别潜在治疗化合物的能力。总之,AlzyFinder 的创新方法通过结合网络药理学分析超越了传统的虚拟筛选,从而提供了对候选药物的系统作用的见解。此功能允许探索配体-蛋白质和蛋白质-蛋白质相互作用及其延伸,从而提供潜在治疗影响的全面视图。作为同类产品中的第一个开放访问平台,AlzyFinder 是 AD 研究社区的宝贵资源,可通过 http://www.alzyfinder-platform.udec.cl 获得支持数据和脚本,可通过 GitHub https://github.com/ramirezlab/AlzyFinder 访问。
更新日期:2024-10-31
中文翻译:

AlzyFinder:用于基于配体的虚拟筛选和网络药理学的机器学习驱动平台
阿尔茨海默病 (AD) 是一种普遍存在的神经退行性疾病,由于其多因素性质和复杂的病理生理学,给药物开发带来了重大挑战。本研究中引入的 AlzyFinder 平台通过为基于平行配体的虚拟筛选和网络药理学提供一个全面的、免费的基于 Web 的工具来应对这些挑战,专门针对超过 85 种与 AD 相关的关键蛋白。这种创新方法旨在加强对潜在多靶标配体的识别和分析,从而加速开发针对 AD 的有效治疗策略。AlzyFinder 平台整合了机器学习模型,以促进基于配体的虚拟筛选过程。这些模型使用 XGBoost 算法构建并通过 Optuna 进行优化,使用强大的方法进行精心训练和验证,以确保高预测准确性。验证包括对活性、非活性和诱饵分子进行的广泛测试,证明了该平台在区分活性化合物方面的功效。这些模型根据平衡的准确度、精度和 F1 分数指标进行评估。采用独特的软投票集成方法来优化分类过程,整合各个模型的优势。该方法框架能够对相互作用数据进行全面分析,这些数据以多种格式呈现,例如表格、热图和交互式配体-蛋白质相互作用网络,从而增强药物-蛋白质相互作用的可视化和分析。AlzyFinder 用于筛选最近报道的 5 种分子(未用于训练或验证 ML 模型)作为针对 5 个关键 AD 靶标的活性化合物。 该平台通过准确预测所有五个分子为真阳性(概率大于 0.70)来证明其疗效。这一结果强调了该平台高精度识别潜在治疗化合物的能力。总之,AlzyFinder 的创新方法通过结合网络药理学分析超越了传统的虚拟筛选,从而提供了对候选药物的系统作用的见解。此功能允许探索配体-蛋白质和蛋白质-蛋白质相互作用及其延伸,从而提供潜在治疗影响的全面视图。作为同类产品中的第一个开放访问平台,AlzyFinder 是 AD 研究社区的宝贵资源,可通过 http://www.alzyfinder-platform.udec.cl 获得支持数据和脚本,可通过 GitHub https://github.com/ramirezlab/AlzyFinder 访问。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号