当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Stepwise Simulation of 3,5-Dihydro-5-methylidene-4H-imidazol-4-one (MIO) Biogenesis in Histidine Ammonia-lyase
Biochemistry ( IF 2.9 ) Pub Date : 2016-10-10 00:00:00 , DOI: 10.1021/acs.biochem.6b00744 Pedro A. Sánchez-Murcia 1 , Juan A. Bueren-Calabuig 1 , Marta Camacho-Artacho 2 , Álvaro Cortés-Cabrera 1 , Federico Gago 1
Biochemistry ( IF 2.9 ) Pub Date : 2016-10-10 00:00:00 , DOI: 10.1021/acs.biochem.6b00744 Pedro A. Sánchez-Murcia 1 , Juan A. Bueren-Calabuig 1 , Marta Camacho-Artacho 2 , Álvaro Cortés-Cabrera 1 , Federico Gago 1
Affiliation
|
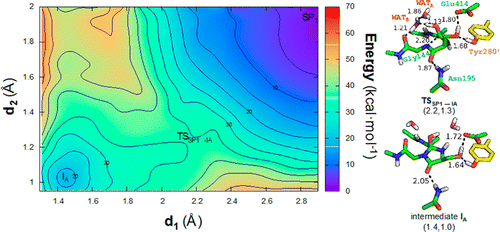
A 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO) electrophilic moiety is post-translationally and autocatalytically generated in homotetrameric histidine ammonia-lyase (HAL) and other enzymes containing the tripeptide Ala-Ser-Gly in a suitably positioned loop. The backbone cyclization step is identical to that taking place during fluorophore formation in green fluorescent protein from the tripeptide Ser-Tyr-Gly, but dehydration, rather than dehydrogenation by molecular oxygen, is the reaction that gives rise to the mature MIO ring system. To gain additional insight into this unique process and shed light on some still unresolved issues, we have made use of extensive molecular dynamics simulations and hybrid quantum mechanics/molecular mechanics calculations implementing the self-consistent charge density functional tight-binding method on a fully solvated tetramer of Pseudomonas putida HAL. Our results strongly support the idea that mechanical compression of the reacting loop by neighboring protein residues in the precursor state is absolutely required to prevent formation of inhibitory main-chain hydrogen bonds and to enforce proper alignment of donor and acceptor orbitals for bond creation. The consideration of the protein environment in our computations shows that water molecules, which have been mostly neglected in previous theoretical work, play a highly relevant role in the reaction mechanism and, more importantly, that backbone cyclization resulting from the nucleophilic attack of the Gly amide lone pair on the π* orbital of the Ala carbonyl precedes side-chain dehydration of the central serine.
中文翻译:

组氨酸氨裂解酶中3,5-二氢-5-亚甲基-4 H-咪唑-4-酮(MIO)生物发生的逐步模拟
3,5-二氢-5-亚甲基-4 H-在同四聚体组氨酸氨裂合酶(HAL)和其他包含三肽Ala-Ser-Gly的酶中,在适当定位的环中翻译后和自催化生成咪唑-4-酮(MIO)亲电子部分。主链环化步骤与三肽Ser-Tyr-Gly在绿色荧光蛋白中形成荧光团的过程相同,但是脱水而不是分子氧脱氢是产生成熟MIO环系统的反应。为了获得对该独特过程的更多了解并阐明一些仍未解决的问题,我们利用了广泛的分子动力学模拟和混合量子力学/分子力学计算,在完全溶剂化的情况下实现了自洽电荷密度功能紧密结合方法的四聚体恶臭假单胞菌HAL。我们的结果强烈支持这样的想法,即绝对需要前体状态的相邻蛋白质残基对反应环进行机械压缩,以防止形成抑制性主链氢键并强制供体和受体轨道正确对齐以形成键。在我们的计算中对蛋白质环境的考虑表明,在先前的理论工作中大多被忽略的水分子在反应机理中起着非常重要的作用,更重要的是,甘氨酸酰胺的亲核攻击导致主链环化丙氨酸Ala的π*轨道上的孤对在中央丝氨酸的侧链脱水之前。
更新日期:2016-10-10
中文翻译:

组氨酸氨裂解酶中3,5-二氢-5-亚甲基-4 H-咪唑-4-酮(MIO)生物发生的逐步模拟
3,5-二氢-5-亚甲基-4 H-在同四聚体组氨酸氨裂合酶(HAL)和其他包含三肽Ala-Ser-Gly的酶中,在适当定位的环中翻译后和自催化生成咪唑-4-酮(MIO)亲电子部分。主链环化步骤与三肽Ser-Tyr-Gly在绿色荧光蛋白中形成荧光团的过程相同,但是脱水而不是分子氧脱氢是产生成熟MIO环系统的反应。为了获得对该独特过程的更多了解并阐明一些仍未解决的问题,我们利用了广泛的分子动力学模拟和混合量子力学/分子力学计算,在完全溶剂化的情况下实现了自洽电荷密度功能紧密结合方法的四聚体恶臭假单胞菌HAL。我们的结果强烈支持这样的想法,即绝对需要前体状态的相邻蛋白质残基对反应环进行机械压缩,以防止形成抑制性主链氢键并强制供体和受体轨道正确对齐以形成键。在我们的计算中对蛋白质环境的考虑表明,在先前的理论工作中大多被忽略的水分子在反应机理中起着非常重要的作用,更重要的是,甘氨酸酰胺的亲核攻击导致主链环化丙氨酸Ala的π*轨道上的孤对在中央丝氨酸的侧链脱水之前。











































 京公网安备 11010802027423号
京公网安备 11010802027423号